Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
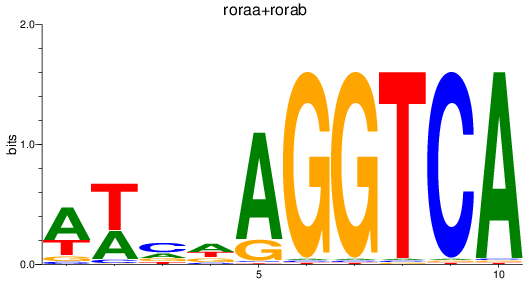
Results for roraa+rorab
Z-value: 0.71
Transcription factors associated with roraa+rorab
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rorab
|
ENSDARG00000001910 | RAR-related orphan receptor A, paralog b |
|
roraa
|
ENSDARG00000031768 | RAR-related orphan receptor A, paralog a |
|
rorab
|
ENSDARG00000109324 | RAR-related orphan receptor A, paralog b |
|
rorab
|
ENSDARG00000113275 | RAR-related orphan receptor A, paralog b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rorab | dr11_v1_chr7_-_29571615_29571615 | 0.64 | 3.2e-03 | Click! |
| roraa | dr11_v1_chr25_+_33849647_33849647 | 0.60 | 6.8e-03 | Click! |
Activity profile of roraa+rorab motif
Sorted Z-values of roraa+rorab motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_11385155 | 2.69 |
ENSDART00000064214
|
plac8.1
|
placenta-specific 8, tandem duplicate 1 |
| chr19_+_40856534 | 2.04 |
ENSDART00000051950
|
gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr13_+_50375800 | 2.03 |
ENSDART00000099537
|
cox5b2
|
cytochrome c oxidase subunit Vb 2 |
| chr19_+_40856807 | 1.89 |
ENSDART00000139083
|
gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr15_+_19797918 | 1.66 |
ENSDART00000113314
|
si:ch211-229d2.5
|
si:ch211-229d2.5 |
| chr25_+_3326885 | 1.55 |
ENSDART00000104866
|
ldhbb
|
lactate dehydrogenase Bb |
| chr15_-_12270857 | 1.53 |
ENSDART00000170093
|
si:dkey-36i7.3
|
si:dkey-36i7.3 |
| chr1_-_50859053 | 1.40 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr11_+_30729745 | 1.39 |
ENSDART00000103270
|
slc22a7a
|
solute carrier family 22 (organic anion transporter), member 7a |
| chr6_-_35439406 | 1.37 |
ENSDART00000073784
|
rgs5a
|
regulator of G protein signaling 5a |
| chr23_+_19564392 | 1.30 |
ENSDART00000144746
|
atp6ap1lb
|
ATPase H+ transporting accessory protein 1 like b |
| chr5_-_41831646 | 1.27 |
ENSDART00000134326
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr18_+_8340886 | 1.18 |
ENSDART00000081132
|
cpt1b
|
carnitine palmitoyltransferase 1B (muscle) |
| chr7_+_29133321 | 1.17 |
ENSDART00000052346
|
gnao1b
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O, b |
| chr21_+_7582036 | 1.14 |
ENSDART00000135485
ENSDART00000027268 |
otpa
|
orthopedia homeobox a |
| chr14_-_33454595 | 1.08 |
ENSDART00000109615
ENSDART00000173267 ENSDART00000185737 ENSDART00000190989 |
tmem255a
|
transmembrane protein 255A |
| chr5_-_68916455 | 1.06 |
ENSDART00000171465
|
ank1a
|
ankyrin 1, erythrocytic a |
| chr25_+_3327071 | 1.05 |
ENSDART00000136131
ENSDART00000133243 |
ldhbb
|
lactate dehydrogenase Bb |
| chr6_-_28980756 | 1.05 |
ENSDART00000014661
|
glmnb
|
glomulin, FKBP associated protein b |
| chr20_+_5564042 | 1.02 |
ENSDART00000090934
ENSDART00000127050 |
nrxn3b
|
neurexin 3b |
| chr7_-_48805181 | 0.97 |
ENSDART00000015884
|
mfge8a
|
milk fat globule-EGF factor 8 protein a |
| chr10_-_17103651 | 0.90 |
ENSDART00000108959
|
RNF208
|
ring finger protein 208 |
| chr20_+_16750177 | 0.88 |
ENSDART00000185357
|
calm1b
|
calmodulin 1b |
| chr19_+_19976990 | 0.79 |
ENSDART00000052627
|
npvf
|
neuropeptide VF precursor |
| chr17_-_7818944 | 0.78 |
ENSDART00000135538
ENSDART00000037541 |
rmnd1
|
required for meiotic nuclear division 1 homolog |
| chr10_-_29900546 | 0.74 |
ENSDART00000147441
|
lim2.1
|
lens intrinsic membrane protein 2.1 |
| chr16_-_28856112 | 0.72 |
ENSDART00000078543
|
syt11b
|
synaptotagmin XIb |
| chr21_+_5882300 | 0.72 |
ENSDART00000165065
|
uqcr10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr6_-_39764995 | 0.70 |
ENSDART00000085277
|
pfkmb
|
phosphofructokinase, muscle b |
| chr1_-_45177373 | 0.68 |
ENSDART00000143142
ENSDART00000034549 |
zgc:111983
|
zgc:111983 |
| chr25_+_31264155 | 0.68 |
ENSDART00000012256
|
tnni2a.3
|
troponin I type 2a (skeletal, fast), tandem duplicate 3 |
| chr20_-_14680897 | 0.66 |
ENSDART00000063857
ENSDART00000161314 |
scrn2
|
secernin 2 |
| chr4_-_1360495 | 0.65 |
ENSDART00000164623
|
ptn
|
pleiotrophin |
| chr15_+_45575180 | 0.63 |
ENSDART00000180441
|
cldn15lb
|
claudin 15-like b |
| chr19_-_5103313 | 0.63 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr19_-_5805923 | 0.62 |
ENSDART00000134340
|
si:ch211-264f5.8
|
si:ch211-264f5.8 |
| chr17_+_33340675 | 0.62 |
ENSDART00000184396
ENSDART00000077553 |
xdh
|
xanthine dehydrogenase |
| chr19_-_5103141 | 0.62 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr23_+_44741500 | 0.61 |
ENSDART00000166421
|
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr12_-_47648538 | 0.60 |
ENSDART00000108477
|
fh
|
fumarate hydratase |
| chr18_+_16330025 | 0.60 |
ENSDART00000142353
|
nts
|
neurotensin |
| chr10_+_17850934 | 0.59 |
ENSDART00000113666
ENSDART00000145936 |
phf24
|
PHD finger protein 24 |
| chr4_+_2620751 | 0.59 |
ENSDART00000013924
|
gpr22a
|
G protein-coupled receptor 22a |
| chr10_-_29101715 | 0.58 |
ENSDART00000149674
ENSDART00000171194 ENSDART00000192019 |
si:ch211-103f14.3
|
si:ch211-103f14.3 |
| chr24_+_29449690 | 0.57 |
ENSDART00000105743
ENSDART00000193556 ENSDART00000145816 |
ntng1a
|
netrin g1a |
| chr10_+_20113830 | 0.57 |
ENSDART00000139722
|
dmtn
|
dematin actin binding protein |
| chr5_-_41838354 | 0.57 |
ENSDART00000146793
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr6_-_49873020 | 0.57 |
ENSDART00000148511
|
gnas
|
GNAS complex locus |
| chr16_+_12236339 | 0.56 |
ENSDART00000132468
|
tpi1b
|
triosephosphate isomerase 1b |
| chr5_+_31860043 | 0.55 |
ENSDART00000036235
ENSDART00000140541 |
iscub
|
iron-sulfur cluster assembly enzyme b |
| chr17_+_25290136 | 0.54 |
ENSDART00000173295
|
kbtbd11
|
kelch repeat and BTB (POZ) domain containing 11 |
| chr2_-_4797512 | 0.54 |
ENSDART00000160765
|
tnk2b
|
tyrosine kinase, non-receptor, 2b |
| chr6_-_10835849 | 0.54 |
ENSDART00000005903
ENSDART00000135065 |
atp5mc3b
|
ATP synthase membrane subunit c locus 3b |
| chr7_+_50109239 | 0.52 |
ENSDART00000021605
|
LRRC4C (1 of many)
|
si:dkey-6l15.1 |
| chr12_+_46708920 | 0.52 |
ENSDART00000153089
|
exoc7
|
exocyst complex component 7 |
| chr7_-_24046999 | 0.52 |
ENSDART00000144616
ENSDART00000124653 ENSDART00000127813 |
dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr3_-_26109322 | 0.52 |
ENSDART00000113780
|
zgc:162612
|
zgc:162612 |
| chr5_+_5689476 | 0.51 |
ENSDART00000022729
|
unm_sa808
|
un-named sa808 |
| chr1_+_19515228 | 0.49 |
ENSDART00000103091
|
clrn2
|
clarin 2 |
| chr11_+_25278772 | 0.48 |
ENSDART00000188630
|
cyldb
|
cylindromatosis (turban tumor syndrome), b |
| chr3_-_19091024 | 0.48 |
ENSDART00000188485
ENSDART00000110554 |
grin2ca
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2Ca |
| chr17_+_30448452 | 0.48 |
ENSDART00000153939
|
lpin1
|
lipin 1 |
| chr2_+_30787128 | 0.48 |
ENSDART00000189233
|
atp6v1h
|
ATPase H+ transporting V1 subunit H |
| chr21_-_22724980 | 0.48 |
ENSDART00000035469
|
c1qa
|
complement component 1, q subcomponent, A chain |
| chr3_+_1182315 | 0.46 |
ENSDART00000055430
|
ndufa6
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 6 |
| chr5_+_1219187 | 0.45 |
ENSDART00000129490
|
gb:bc139872
|
expressed sequence BC139872 |
| chr2_-_56655769 | 0.45 |
ENSDART00000113589
|
gpx4b
|
glutathione peroxidase 4b |
| chr7_-_29341233 | 0.44 |
ENSDART00000140938
ENSDART00000147251 |
trpm1a
|
transient receptor potential cation channel, subfamily M, member 1a |
| chr10_+_19528321 | 0.43 |
ENSDART00000184816
|
vsig8a
|
V-set and immunoglobulin domain containing 8a |
| chr1_+_9708801 | 0.42 |
ENSDART00000189621
|
elfn1b
|
extracellular leucine-rich repeat and fibronectin type III domain containing 1b |
| chr15_+_1705167 | 0.42 |
ENSDART00000081940
|
otol1b
|
otolin 1b |
| chr11_+_28476298 | 0.41 |
ENSDART00000122319
|
lrrc38b
|
leucine rich repeat containing 38b |
| chr17_+_22381215 | 0.41 |
ENSDART00000162670
ENSDART00000128875 |
slc8a1b
|
solute carrier family 8 (sodium/calcium exchanger), member 1b |
| chr15_+_29362714 | 0.40 |
ENSDART00000099916
ENSDART00000126196 |
gucy2f
|
guanylate cyclase 2F, retinal |
| chr10_-_25561751 | 0.40 |
ENSDART00000147089
|
grik1a
|
glutamate receptor, ionotropic, kainate 1a |
| chr3_-_26191960 | 0.40 |
ENSDART00000113843
|
ypel3
|
yippee-like 3 |
| chr19_-_5332784 | 0.39 |
ENSDART00000010373
|
krt1-19d
|
keratin, type 1, gene 19d |
| chr20_+_40150612 | 0.39 |
ENSDART00000143680
ENSDART00000109681 ENSDART00000101041 ENSDART00000121818 |
trdn
|
triadin |
| chr9_-_16877456 | 0.39 |
ENSDART00000161105
ENSDART00000160869 |
fbxl3a
|
F-box and leucine-rich repeat protein 3a |
| chr2_+_34767171 | 0.39 |
ENSDART00000145451
|
astn1
|
astrotactin 1 |
| chr12_-_45876387 | 0.38 |
ENSDART00000043210
ENSDART00000149044 |
pax2b
|
paired box 2b |
| chr18_-_42333428 | 0.38 |
ENSDART00000034225
|
cntn5
|
contactin 5 |
| chr19_-_32487469 | 0.38 |
ENSDART00000050130
|
gmpr
|
guanosine monophosphate reductase |
| chr1_-_44704261 | 0.38 |
ENSDART00000133210
|
si:dkey-28b4.8
|
si:dkey-28b4.8 |
| chr20_-_25709247 | 0.37 |
ENSDART00000146711
|
si:dkeyp-117h8.2
|
si:dkeyp-117h8.2 |
| chr1_+_44826593 | 0.36 |
ENSDART00000162200
|
STX3
|
zgc:165520 |
| chr17_-_45254585 | 0.36 |
ENSDART00000185507
ENSDART00000172080 |
ttbk2a
|
tau tubulin kinase 2a |
| chr18_-_1185772 | 0.36 |
ENSDART00000143245
|
nptnb
|
neuroplastin b |
| chr18_-_42313798 | 0.35 |
ENSDART00000098639
|
cntn5
|
contactin 5 |
| chr13_-_31452516 | 0.35 |
ENSDART00000193268
|
rtn1a
|
reticulon 1a |
| chr14_+_33882973 | 0.34 |
ENSDART00000019396
|
clic2
|
chloride intracellular channel 2 |
| chr13_-_29406534 | 0.34 |
ENSDART00000100877
|
zgc:153142
|
zgc:153142 |
| chr6_-_39765546 | 0.34 |
ENSDART00000185767
|
pfkmb
|
phosphofructokinase, muscle b |
| chr7_-_16205471 | 0.34 |
ENSDART00000173584
|
btr05
|
bloodthirsty-related gene family, member 5 |
| chr25_-_31396479 | 0.33 |
ENSDART00000156828
|
prr33
|
proline rich 33 |
| chr24_+_39186940 | 0.33 |
ENSDART00000155817
|
spsb3b
|
splA/ryanodine receptor domain and SOCS box containing 3b |
| chr14_-_30490465 | 0.33 |
ENSDART00000173107
|
micu3b
|
mitochondrial calcium uptake family, member 3b |
| chr10_+_40973524 | 0.32 |
ENSDART00000176660
ENSDART00000114966 |
antxr1b
|
anthrax toxin receptor 1b |
| chr19_-_3574060 | 0.32 |
ENSDART00000105120
|
tmem170b
|
transmembrane protein 170B |
| chr22_-_11124419 | 0.32 |
ENSDART00000149634
|
atp6ap2
|
ATPase H+ transporting accessory protein 2 |
| chr5_-_55395964 | 0.32 |
ENSDART00000145791
|
prune2
|
prune homolog 2 (Drosophila) |
| chr2_-_16380283 | 0.32 |
ENSDART00000149992
|
si:dkey-231j24.3
|
si:dkey-231j24.3 |
| chr4_+_20954929 | 0.32 |
ENSDART00000143674
|
nav3
|
neuron navigator 3 |
| chr19_+_30990815 | 0.32 |
ENSDART00000134645
|
sync
|
syncoilin, intermediate filament protein |
| chr2_+_10127762 | 0.31 |
ENSDART00000100726
|
insl5b
|
insulin-like 5b |
| chr7_-_4036184 | 0.31 |
ENSDART00000019949
|
ndrg2
|
NDRG family member 2 |
| chr6_+_51773873 | 0.30 |
ENSDART00000156516
|
tmem74b
|
transmembrane protein 74B |
| chr11_-_37995501 | 0.30 |
ENSDART00000192096
|
slc41a1
|
solute carrier family 41 (magnesium transporter), member 1 |
| chr18_+_45571378 | 0.30 |
ENSDART00000077251
|
kifc3
|
kinesin family member C3 |
| chr1_+_34496855 | 0.30 |
ENSDART00000012873
|
klf12a
|
Kruppel-like factor 12a |
| chr5_-_41841892 | 0.29 |
ENSDART00000167089
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr19_+_30662529 | 0.29 |
ENSDART00000175662
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr23_+_42813415 | 0.28 |
ENSDART00000055577
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr7_-_56180680 | 0.28 |
ENSDART00000098438
|
spg7
|
spastic paraplegia 7 |
| chr7_+_67494107 | 0.27 |
ENSDART00000185653
|
cpne7
|
copine VII |
| chr12_-_45875946 | 0.27 |
ENSDART00000149970
|
pax2b
|
paired box 2b |
| chr2_-_22659450 | 0.27 |
ENSDART00000115025
|
thap4
|
THAP domain containing 4 |
| chr5_+_57658898 | 0.27 |
ENSDART00000074268
ENSDART00000124568 |
zgc:153929
|
zgc:153929 |
| chr3_+_49074008 | 0.26 |
ENSDART00000168864
|
zgc:112146
|
zgc:112146 |
| chr23_-_19953089 | 0.26 |
ENSDART00000153828
|
atp2b3b
|
ATPase plasma membrane Ca2+ transporting 3b |
| chr4_-_13502549 | 0.26 |
ENSDART00000140366
|
si:ch211-266a5.12
|
si:ch211-266a5.12 |
| chr19_+_30990129 | 0.25 |
ENSDART00000052169
ENSDART00000193376 |
sync
|
syncoilin, intermediate filament protein |
| chr6_+_45692026 | 0.25 |
ENSDART00000164759
|
cntn4
|
contactin 4 |
| chr16_+_26738404 | 0.25 |
ENSDART00000163110
|
fsbp
|
fibrinogen silencer binding protein |
| chr18_+_7594012 | 0.24 |
ENSDART00000062150
|
zgc:77752
|
zgc:77752 |
| chr9_-_30808073 | 0.23 |
ENSDART00000146300
|
klf5b
|
Kruppel-like factor 5b |
| chr6_+_40523370 | 0.23 |
ENSDART00000033819
|
prkcda
|
protein kinase C, delta a |
| chr7_+_44484853 | 0.23 |
ENSDART00000189079
ENSDART00000121826 |
bean1
|
brain expressed, associated with NEDD4, 1 |
| chr22_+_29135846 | 0.22 |
ENSDART00000143811
|
baiap2l2b
|
BAI1-associated protein 2-like 2b |
| chr14_+_33525196 | 0.21 |
ENSDART00000085335
|
zdhhc9
|
zinc finger, DHHC-type containing 9 |
| chr17_-_12718547 | 0.21 |
ENSDART00000193002
|
BX294375.1
|
|
| chr3_+_12861158 | 0.21 |
ENSDART00000171237
|
kcnj2b
|
potassium inwardly-rectifying channel, subfamily J, member 2b |
| chr16_+_27349585 | 0.21 |
ENSDART00000142573
|
nr4a3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr3_-_60142530 | 0.21 |
ENSDART00000153247
|
si:ch211-120g10.1
|
si:ch211-120g10.1 |
| chr25_-_19374710 | 0.20 |
ENSDART00000184483
ENSDART00000188706 |
map1ab
|
microtubule-associated protein 1Ab |
| chr16_-_16619854 | 0.19 |
ENSDART00000150512
ENSDART00000191306 ENSDART00000181773 ENSDART00000183231 |
cyp21a2
|
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chr14_+_2095394 | 0.19 |
ENSDART00000186847
|
CT573264.2
|
|
| chr10_+_1638876 | 0.19 |
ENSDART00000184484
ENSDART00000060946 ENSDART00000181251 |
sgsm1b
|
small G protein signaling modulator 1b |
| chr2_-_127945 | 0.18 |
ENSDART00000056453
|
igfbp1b
|
insulin-like growth factor binding protein 1b |
| chr3_-_39198113 | 0.17 |
ENSDART00000102690
|
RETSAT
|
zgc:154169 |
| chr15_-_30450898 | 0.17 |
ENSDART00000156584
|
msi2b
|
musashi RNA-binding protein 2b |
| chr13_+_33688474 | 0.17 |
ENSDART00000161465
|
CABZ01087953.1
|
|
| chr8_-_50132860 | 0.17 |
ENSDART00000149964
|
antxr1a
|
anthrax toxin receptor 1a |
| chr4_-_17629444 | 0.17 |
ENSDART00000108814
|
nrip2
|
nuclear receptor interacting protein 2 |
| chr1_+_12763920 | 0.17 |
ENSDART00000189465
|
pcdh10a
|
protocadherin 10a |
| chr15_-_6966221 | 0.17 |
ENSDART00000165487
ENSDART00000027657 |
mrps22
|
mitochondrial ribosomal protein S22 |
| chr12_+_46634736 | 0.16 |
ENSDART00000008009
|
trim16
|
tripartite motif containing 16 |
| chr10_-_32524771 | 0.16 |
ENSDART00000066793
|
dgat2
|
diacylglycerol O-acyltransferase 2 |
| chr12_-_44199316 | 0.15 |
ENSDART00000170378
|
si:ch73-329n5.1
|
si:ch73-329n5.1 |
| chr11_+_5842632 | 0.15 |
ENSDART00000111374
ENSDART00000158599 |
ndufs7
|
NADH dehydrogenase (ubiquinone) Fe-S protein 7, (NADH-coenzyme Q reductase) |
| chr21_+_20771082 | 0.15 |
ENSDART00000079732
|
oxct1b
|
3-oxoacid CoA transferase 1b |
| chr11_-_29650930 | 0.15 |
ENSDART00000166969
|
chd5
|
chromodomain helicase DNA binding protein 5 |
| chr1_+_59314675 | 0.14 |
ENSDART00000161872
ENSDART00000160658 ENSDART00000169792 ENSDART00000160735 |
parn
|
poly(A)-specific ribonuclease (deadenylation nuclease) |
| chr16_-_39195318 | 0.14 |
ENSDART00000058546
|
ebag9
|
estrogen receptor binding site associated, antigen, 9 |
| chr21_+_11885404 | 0.14 |
ENSDART00000092015
|
dcaf12
|
DDB1 and CUL4 associated factor 12 |
| chr19_+_20724347 | 0.13 |
ENSDART00000090757
|
kat2b
|
K(lysine) acetyltransferase 2B |
| chr18_-_14922872 | 0.13 |
ENSDART00000091722
|
panx2
|
pannexin 2 |
| chr10_+_4924388 | 0.13 |
ENSDART00000108595
|
slc46a2
|
solute carrier family 46 member 2 |
| chr8_+_21225064 | 0.12 |
ENSDART00000129210
|
cry1ba
|
cryptochrome circadian clock 1ba |
| chr10_+_20128267 | 0.12 |
ENSDART00000064615
|
dmtn
|
dematin actin binding protein |
| chr3_+_58167288 | 0.12 |
ENSDART00000155874
ENSDART00000010395 |
uqcrc2a
|
ubiquinol-cytochrome c reductase core protein 2a |
| chr9_-_21067971 | 0.12 |
ENSDART00000004333
|
tbx15
|
T-box 15 |
| chr8_+_2530065 | 0.11 |
ENSDART00000063943
|
mrpl40
|
mitochondrial ribosomal protein L40 |
| chr8_+_18464235 | 0.11 |
ENSDART00000110571
|
alkal1
|
ALK and LTK ligand 1 |
| chr5_+_36870737 | 0.11 |
ENSDART00000145182
|
slc8a2a
|
solute carrier family 8 (sodium/calcium exchanger), member 2a |
| chr13_-_22907260 | 0.10 |
ENSDART00000143097
|
rufy2
|
RUN and FYVE domain containing 2 |
| chr19_+_2590182 | 0.10 |
ENSDART00000162293
|
si:ch73-345f18.3
|
si:ch73-345f18.3 |
| chr22_+_20546612 | 0.10 |
ENSDART00000141852
|
si:dkey-172o19.2
|
si:dkey-172o19.2 |
| chr19_+_10331325 | 0.10 |
ENSDART00000143930
|
tmem238a
|
transmembrane protein 238a |
| chr16_+_41517188 | 0.10 |
ENSDART00000049976
|
si:dkey-11p23.7
|
si:dkey-11p23.7 |
| chr11_+_24900123 | 0.10 |
ENSDART00000044987
ENSDART00000148023 |
timm17a
|
translocase of inner mitochondrial membrane 17 homolog A (yeast) |
| chr14_+_9583588 | 0.09 |
ENSDART00000164101
|
tmem129
|
transmembrane protein 129, E3 ubiquitin protein ligase |
| chr13_+_31172833 | 0.09 |
ENSDART00000176378
|
CR931802.3
|
|
| chr8_+_25902170 | 0.08 |
ENSDART00000193130
|
rhoab
|
ras homolog gene family, member Ab |
| chr6_-_49159207 | 0.08 |
ENSDART00000041942
|
tspan2a
|
tetraspanin 2a |
| chr6_-_15603675 | 0.08 |
ENSDART00000143502
|
lrrfip1b
|
leucine rich repeat (in FLII) interacting protein 1b |
| chr4_-_25181552 | 0.07 |
ENSDART00000066930
|
atp5f1c
|
ATP synthase F1 subunit gamma |
| chr6_-_52788213 | 0.07 |
ENSDART00000179880
|
rbpjl
|
recombination signal binding protein for immunoglobulin kappa J region-like |
| chr17_+_2214283 | 0.07 |
ENSDART00000189187
|
pak6a
|
p21 protein (Cdc42/Rac)-activated kinase 6a |
| chr7_-_16237861 | 0.06 |
ENSDART00000173647
|
btr07
|
bloodthirsty-related gene family, member 7 |
| chr17_-_7371564 | 0.06 |
ENSDART00000060336
|
rab32b
|
RAB32b, member RAS oncogene family |
| chr3_+_32698424 | 0.06 |
ENSDART00000055340
|
fus
|
FUS RNA binding protein |
| chr9_+_4306122 | 0.06 |
ENSDART00000193722
ENSDART00000190521 |
kalrna
|
kalirin RhoGEF kinase a |
| chr8_+_18463797 | 0.06 |
ENSDART00000148958
|
alkal1
|
ALK and LTK ligand 1 |
| chr9_+_22359919 | 0.06 |
ENSDART00000009591
|
crygs4
|
crystallin, gamma S4 |
| chr17_-_36513936 | 0.06 |
ENSDART00000165075
|
DCDC2C
|
si:dkey-25g12.4 |
| chr25_-_1235457 | 0.05 |
ENSDART00000093093
|
coro2bb
|
coronin, actin binding protein, 2Bb |
| chr12_-_25201576 | 0.05 |
ENSDART00000077188
|
cox7a3
|
cytochrome c oxidase subunit VIIa polypeptide 3 |
| chr9_+_23224761 | 0.05 |
ENSDART00000142008
|
map3k19
|
mitogen-activated protein kinase kinase kinase 19 |
| chr8_+_51084993 | 0.05 |
ENSDART00000127709
ENSDART00000053768 |
DISP3
|
si:dkey-32e23.6 |
| chr7_+_39079071 | 0.05 |
ENSDART00000181845
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr23_-_5683147 | 0.04 |
ENSDART00000102766
ENSDART00000067351 |
tnnt2a
|
troponin T type 2a (cardiac) |
| chr15_+_34675660 | 0.04 |
ENSDART00000009127
|
lta
|
lymphotoxin alpha (TNF superfamily, member 1) |
| chr16_-_11789108 | 0.04 |
ENSDART00000060142
|
tlr21
|
toll-like receptor 21 |
| chr12_+_28799988 | 0.04 |
ENSDART00000022724
|
pnpo
|
pyridoxamine 5'-phosphate oxidase |
| chr8_+_14792830 | 0.04 |
ENSDART00000139972
|
cacna1ea
|
calcium channel, voltage-dependent, R type, alpha 1E subunit a |
| chr10_-_26273629 | 0.04 |
ENSDART00000147790
|
dchs1b
|
dachsous cadherin-related 1b |
| chr16_-_50229193 | 0.04 |
ENSDART00000161782
ENSDART00000010081 |
etfb
|
electron-transfer-flavoprotein, beta polypeptide |
| chr2_+_2169337 | 0.04 |
ENSDART00000179939
|
higd1a
|
HIG1 hypoxia inducible domain family, member 1A |
| chr22_+_19486711 | 0.04 |
ENSDART00000144389
ENSDART00000139256 ENSDART00000160855 |
si:dkey-78l4.10
|
si:dkey-78l4.10 |
Network of associatons between targets according to the STRING database.
First level regulatory network of roraa+rorab
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.2 | 0.6 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.2 | 0.8 | GO:0060986 | regulation of endocrine process(GO:0044060) positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) endocrine hormone secretion(GO:0060986) |
| 0.2 | 0.8 | GO:0070131 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.2 | 1.1 | GO:0035176 | mammillary body development(GO:0021767) social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.2 | 2.0 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.6 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.1 | 1.0 | GO:0006735 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 0.7 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 0.4 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.1 | 0.5 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.1 | 0.6 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.5 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.1 | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) electron transport coupled proton transport(GO:0015990) |
| 0.0 | 0.5 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.0 | 0.1 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.0 | 0.7 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 0.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 1.0 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.6 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 1.2 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 1.9 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.3 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 0.7 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.2 | GO:0070375 | ERK5 cascade(GO:0070375) regulation of ERK5 cascade(GO:0070376) positive regulation of ERK5 cascade(GO:0070378) |
| 0.0 | 0.2 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.5 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.0 | 0.5 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.3 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.0 | 0.6 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.1 | GO:0045124 | regulation of bone resorption(GO:0045124) |
| 0.0 | 0.5 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.3 | GO:0072576 | liver morphogenesis(GO:0072576) |
| 0.0 | 0.2 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.7 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.6 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.0 | 0.4 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.6 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.1 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.0 | 0.3 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.4 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.0 | 1.0 | GO:0007338 | single fertilization(GO:0007338) |
| 0.0 | 0.4 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.2 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.5 | GO:0046460 | triglyceride biosynthetic process(GO:0019432) neutral lipid biosynthetic process(GO:0046460) acylglycerol biosynthetic process(GO:0046463) |
| 0.0 | 0.5 | GO:0006958 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.0 | GO:0046184 | aldehyde biosynthetic process(GO:0046184) |
| 0.0 | 0.4 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.4 | GO:0060401 | cytosolic calcium ion transport(GO:0060401) calcium ion transport into cytosol(GO:0060402) |
| 0.0 | 0.7 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 1.1 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.1 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.3 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.0 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.0 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.0 | 0.2 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.7 | GO:0051781 | positive regulation of cell division(GO:0051781) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.9 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.2 | 1.3 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.1 | 0.4 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 1.0 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.7 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 0.4 | GO:0098651 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.1 | 0.5 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.1 | 0.6 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 1.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.5 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.6 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.3 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.4 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.0 | 0.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.6 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.4 | GO:0035869 | ciliary transition zone(GO:0035869) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.4 | 2.6 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.3 | 1.0 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.3 | 1.7 | GO:0051430 | mu-type opioid receptor binding(GO:0031852) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.2 | 3.9 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.7 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.1 | 0.4 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 0.7 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 1.0 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 0.4 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.1 | 2.0 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.6 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.0 | 0.5 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 1.0 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.3 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.6 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.5 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.3 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 0.9 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.2 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.0 | 0.5 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.4 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.5 | GO:0036442 | hydrogen-exporting ATPase activity(GO:0036442) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.6 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 2.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.0 | 0.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.2 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.7 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.5 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.5 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.6 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.3 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.7 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.2 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.5 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.6 | PID AP1 PATHWAY | AP-1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.9 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 0.6 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 1.2 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.5 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.6 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.5 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.1 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |