Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
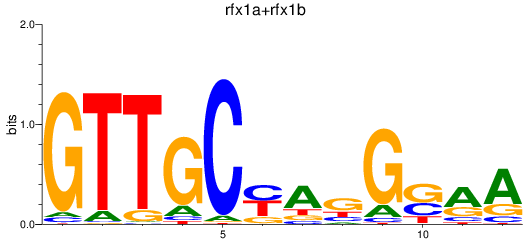
Results for rfx1a+rfx1b
Z-value: 0.92
Transcription factors associated with rfx1a+rfx1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rfx1a
|
ENSDARG00000005883 | regulatory factor X, 1a (influences HLA class II expression) |
|
rfx1b
|
ENSDARG00000075904 | regulatory factor X, 1b (influences HLA class II expression) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rfx1a | dr11_v1_chr3_-_19200571_19200651 | 0.66 | 2.1e-03 | Click! |
| rfx1b | dr11_v1_chr1_-_54107321_54107321 | 0.57 | 1.1e-02 | Click! |
Activity profile of rfx1a+rfx1b motif
Sorted Z-values of rfx1a+rfx1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_52902693 | 3.03 |
ENSDART00000166115
ENSDART00000161050 |
ctsbb
|
cathepsin Bb |
| chr13_-_51952020 | 2.23 |
ENSDART00000181079
|
LT631676.1
|
|
| chr8_-_53488832 | 1.88 |
ENSDART00000191801
|
chdh
|
choline dehydrogenase |
| chr8_-_5220125 | 1.56 |
ENSDART00000035676
|
bnip3la
|
BCL2 interacting protein 3 like a |
| chr21_-_1625976 | 1.38 |
ENSDART00000066621
|
vmo1b
|
vitelline membrane outer layer 1 homolog b |
| chr23_+_10469955 | 1.34 |
ENSDART00000140557
|
tns2a
|
tensin 2a |
| chr23_+_44665230 | 1.26 |
ENSDART00000144958
|
camta2
|
calmodulin binding transcription activator 2 |
| chr24_-_39772045 | 1.19 |
ENSDART00000087441
|
GFOD1
|
si:ch211-276f18.2 |
| chr16_-_9869056 | 1.11 |
ENSDART00000149312
|
ncalda
|
neurocalcin delta a |
| chr25_+_34407740 | 1.09 |
ENSDART00000012677
|
OTUD7A
|
si:dkey-37f18.2 |
| chr3_-_6719232 | 1.06 |
ENSDART00000154294
|
atg4db
|
autophagy related 4D, cysteine peptidase b |
| chr20_+_49787584 | 1.02 |
ENSDART00000193458
ENSDART00000181511 ENSDART00000185850 ENSDART00000185613 ENSDART00000191671 |
CABZ01078261.1
|
|
| chr11_+_43661735 | 0.95 |
ENSDART00000017912
|
gli2b
|
GLI family zinc finger 2b |
| chr5_-_38384289 | 0.94 |
ENSDART00000135260
|
mink1
|
misshapen-like kinase 1 |
| chr3_+_17456428 | 0.93 |
ENSDART00000090676
ENSDART00000182082 |
si:ch211-210g13.5
|
si:ch211-210g13.5 |
| chr19_+_43123331 | 0.90 |
ENSDART00000187836
|
CABZ01101996.1
|
|
| chr3_+_49521106 | 0.90 |
ENSDART00000162799
|
crb3a
|
crumbs homolog 3a |
| chr1_+_50538839 | 0.90 |
ENSDART00000020412
|
pkd2
|
polycystic kidney disease 2 |
| chr13_-_24717365 | 0.90 |
ENSDART00000137934
ENSDART00000003922 |
erlin1
|
ER lipid raft associated 1 |
| chr13_-_18691041 | 0.90 |
ENSDART00000057867
|
sfxn3
|
sideroflexin 3 |
| chr9_-_41401564 | 0.89 |
ENSDART00000059628
|
nab1b
|
NGFI-A binding protein 1b (EGR1 binding protein 1) |
| chr18_+_29950233 | 0.88 |
ENSDART00000146431
|
atmin
|
ATM interactor |
| chr17_+_34186632 | 0.87 |
ENSDART00000014306
|
mpp5a
|
membrane protein, palmitoylated 5a (MAGUK p55 subfamily member 5) |
| chr23_+_21380079 | 0.85 |
ENSDART00000089379
|
iffo2a
|
intermediate filament family orphan 2a |
| chr22_+_438714 | 0.85 |
ENSDART00000136491
|
celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr10_+_6907715 | 0.85 |
ENSDART00000041068
|
slc38a9
|
solute carrier family 38, member 9 |
| chr23_-_12158685 | 0.84 |
ENSDART00000135035
|
fam217b
|
family with sequence similarity 217, member B |
| chr18_-_2727764 | 0.84 |
ENSDART00000160841
|
ARHGEF17
|
si:ch211-248g20.5 |
| chr14_-_33481428 | 0.84 |
ENSDART00000147059
ENSDART00000140001 ENSDART00000124242 ENSDART00000164836 ENSDART00000190104 ENSDART00000186833 ENSDART00000180873 |
lamp2
|
lysosomal-associated membrane protein 2 |
| chr13_+_48482256 | 0.83 |
ENSDART00000053332
|
FOXN2 (1 of many)
|
forkhead box N2 |
| chr17_-_22067451 | 0.82 |
ENSDART00000156872
|
ttbk1b
|
tau tubulin kinase 1b |
| chr1_+_604127 | 0.82 |
ENSDART00000133165
|
jam2a
|
junctional adhesion molecule 2a |
| chr13_-_47556447 | 0.81 |
ENSDART00000131239
|
acoxl
|
acyl-CoA oxidase-like |
| chr6_+_18418651 | 0.81 |
ENSDART00000171097
ENSDART00000167798 |
rab11fip4b
|
RAB11 family interacting protein 4 (class II) b |
| chr21_+_43669943 | 0.77 |
ENSDART00000136025
|
tmlhe
|
trimethyllysine hydroxylase, epsilon |
| chr3_-_46410387 | 0.77 |
ENSDART00000156822
|
cdip1
|
cell death-inducing p53 target 1 |
| chr15_-_43768776 | 0.73 |
ENSDART00000170398
|
grm5b
|
glutamate receptor, metabotropic 5b |
| chr8_-_22838364 | 0.73 |
ENSDART00000187954
ENSDART00000185368 ENSDART00000188210 |
magixa
|
MAGI family member, X-linked a |
| chr20_+_36682051 | 0.73 |
ENSDART00000130513
|
ncoa1
|
nuclear receptor coactivator 1 |
| chr10_-_44482911 | 0.72 |
ENSDART00000085556
|
hip1ra
|
huntingtin interacting protein 1 related a |
| chr24_-_205275 | 0.72 |
ENSDART00000108762
|
vopp1
|
VOPP1, WBP1/VOPP1 family member |
| chr25_+_35942867 | 0.71 |
ENSDART00000066985
|
hsd17b2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr20_-_7583486 | 0.71 |
ENSDART00000144729
|
usp24
|
ubiquitin specific peptidase 24 |
| chr4_-_73520581 | 0.71 |
ENSDART00000171513
|
si:ch73-266f23.1
|
si:ch73-266f23.1 |
| chr22_+_980290 | 0.70 |
ENSDART00000065377
|
def6b
|
differentially expressed in FDCP 6b homolog (mouse) |
| chr17_+_1496107 | 0.70 |
ENSDART00000187804
|
LO018430.1
|
|
| chr20_-_35706379 | 0.70 |
ENSDART00000152847
|
si:dkey-30j22.1
|
si:dkey-30j22.1 |
| chr13_+_13805385 | 0.69 |
ENSDART00000166859
|
slc4a11
|
solute carrier family 4, sodium borate transporter, member 11 |
| chr3_+_49021079 | 0.69 |
ENSDART00000162012
|
zgc:163083
|
zgc:163083 |
| chr20_-_2361226 | 0.69 |
ENSDART00000172130
|
EPB41L2
|
si:ch73-18b11.1 |
| chr17_+_12075805 | 0.68 |
ENSDART00000155329
|
cnsta
|
consortin, connexin sorting protein a |
| chr18_+_50890749 | 0.68 |
ENSDART00000174109
|
si:ch1073-450f2.1
|
si:ch1073-450f2.1 |
| chr19_+_551963 | 0.68 |
ENSDART00000110495
|
akap9
|
A kinase (PRKA) anchor protein 9 |
| chr9_+_48415043 | 0.67 |
ENSDART00000159930
|
lrp2a
|
low density lipoprotein receptor-related protein 2a |
| chr18_+_50880096 | 0.67 |
ENSDART00000169782
|
si:ch1073-450f2.1
|
si:ch1073-450f2.1 |
| chr4_+_15819728 | 0.66 |
ENSDART00000101613
|
lrguk
|
leucine-rich repeats and guanylate kinase domain containing |
| chr17_+_44756247 | 0.65 |
ENSDART00000153773
|
cipca
|
CLOCK-interacting pacemaker a |
| chr15_-_1022436 | 0.65 |
ENSDART00000156003
|
znf1010
|
zinc finger protein 1010 |
| chr22_-_10891213 | 0.64 |
ENSDART00000145229
|
arhgef18b
|
rho/rac guanine nucleotide exchange factor (GEF) 18b |
| chr3_-_12026741 | 0.63 |
ENSDART00000132238
|
cfap70
|
cilia and flagella associated protein 70 |
| chr2_-_1569250 | 0.61 |
ENSDART00000167202
|
dab1b
|
Dab, reelin signal transducer, homolog 1b (Drosophila) |
| chr2_+_55914699 | 0.61 |
ENSDART00000157905
|
atcayb
|
ataxia, cerebellar, Cayman type b |
| chr4_-_9557186 | 0.60 |
ENSDART00000150569
|
shank3b
|
SH3 and multiple ankyrin repeat domains 3b |
| chr8_-_36370552 | 0.58 |
ENSDART00000097932
ENSDART00000148323 |
si:busm1-104n07.3
|
si:busm1-104n07.3 |
| chr13_+_669177 | 0.58 |
ENSDART00000190085
|
CU462915.1
|
|
| chr21_+_3244146 | 0.57 |
ENSDART00000127740
|
ctif
|
CBP80/20-dependent translation initiation factor |
| chr1_+_11711503 | 0.56 |
ENSDART00000138355
|
si:dkey-26i13.8
|
si:dkey-26i13.8 |
| chr8_-_49345388 | 0.56 |
ENSDART00000053203
|
plp2
|
proteolipid protein 2 |
| chr6_+_13933464 | 0.56 |
ENSDART00000109144
|
ptprnb
|
protein tyrosine phosphatase, receptor type, Nb |
| chr18_-_50579629 | 0.55 |
ENSDART00000188252
|
zgc:158464
|
zgc:158464 |
| chr17_+_21964472 | 0.55 |
ENSDART00000063704
ENSDART00000188904 |
crip3
|
cysteine-rich protein 3 |
| chr6_+_612330 | 0.54 |
ENSDART00000166872
ENSDART00000191758 |
kynu
|
kynureninase |
| chr1_+_54683655 | 0.53 |
ENSDART00000132785
|
knop1
|
lysine-rich nucleolar protein 1 |
| chr2_+_7192966 | 0.53 |
ENSDART00000142735
|
si:ch211-13f8.1
|
si:ch211-13f8.1 |
| chr18_-_43884044 | 0.52 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr15_-_47892583 | 0.51 |
ENSDART00000192584
|
DMWD
|
zmp:0000000529 |
| chr6_-_40842768 | 0.51 |
ENSDART00000076160
|
mustn1a
|
musculoskeletal, embryonic nuclear protein 1a |
| chr15_-_47193564 | 0.51 |
ENSDART00000172453
|
LSAMP
|
limbic system-associated membrane protein |
| chr25_+_18436301 | 0.50 |
ENSDART00000056180
|
cep41
|
centrosomal protein 41 |
| chr4_-_9579299 | 0.49 |
ENSDART00000183079
ENSDART00000192968 ENSDART00000091809 |
shank3b
|
SH3 and multiple ankyrin repeat domains 3b |
| chr1_+_19943803 | 0.47 |
ENSDART00000164661
|
apbb2b
|
amyloid beta (A4) precursor protein-binding, family B, member 2b |
| chr2_-_42960353 | 0.46 |
ENSDART00000098303
|
oc90
|
otoconin 90 |
| chr17_-_49412313 | 0.46 |
ENSDART00000152100
|
mthfd1b
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1b |
| chr22_-_36690742 | 0.45 |
ENSDART00000017188
ENSDART00000124698 |
ncl
|
nucleolin |
| chr21_-_32781612 | 0.45 |
ENSDART00000031028
|
cnot6a
|
CCR4-NOT transcription complex, subunit 6a |
| chr17_+_33296852 | 0.45 |
ENSDART00000154580
|
dnajc27
|
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr6_-_37403444 | 0.45 |
ENSDART00000136314
|
cthl
|
cystathionase (cystathionine gamma-lyase), like |
| chr4_+_34343524 | 0.44 |
ENSDART00000171694
|
si:ch211-246b8.5
|
si:ch211-246b8.5 |
| chr23_+_6272638 | 0.44 |
ENSDART00000190366
|
syt2a
|
synaptotagmin IIa |
| chr24_-_1657276 | 0.43 |
ENSDART00000168131
|
si:ch73-378g22.1
|
si:ch73-378g22.1 |
| chr24_+_38155830 | 0.43 |
ENSDART00000152019
|
si:ch211-234p6.5
|
si:ch211-234p6.5 |
| chr8_+_7144066 | 0.43 |
ENSDART00000146306
|
slc6a6a
|
solute carrier family 6 (neurotransmitter transporter), member 6a |
| chr2_-_16449504 | 0.43 |
ENSDART00000144801
|
atr
|
ATR serine/threonine kinase |
| chr12_+_4971515 | 0.43 |
ENSDART00000161076
|
arhgap27
|
Rho GTPase activating protein 27 |
| chr20_-_27857676 | 0.43 |
ENSDART00000192934
|
syndig1l
|
synapse differentiation inducing 1-like |
| chr19_-_43639331 | 0.43 |
ENSDART00000138009
ENSDART00000086138 |
fam83hb
|
family with sequence similarity 83, member Hb |
| chr18_+_49411417 | 0.43 |
ENSDART00000028944
|
LRFN3
|
zmp:0000001073 |
| chr16_+_41717313 | 0.42 |
ENSDART00000015170
ENSDART00000075937 |
mtdhb
|
metadherin b |
| chr5_-_38384755 | 0.42 |
ENSDART00000188573
ENSDART00000051233 |
mink1
|
misshapen-like kinase 1 |
| chr8_-_46045172 | 0.41 |
ENSDART00000135138
|
si:ch211-119d14.3
|
si:ch211-119d14.3 |
| chr2_+_7075220 | 0.41 |
ENSDART00000022963
|
cdc14aa
|
cell division cycle 14Aa |
| chr8_+_36288261 | 0.41 |
ENSDART00000191281
|
CU929676.1
|
|
| chr9_+_48219111 | 0.41 |
ENSDART00000111225
ENSDART00000145972 |
ccdc173
|
coiled-coil domain containing 173 |
| chr3_+_60034194 | 0.40 |
ENSDART00000189858
ENSDART00000153642 |
CR352258.1
|
|
| chr10_-_74408 | 0.40 |
ENSDART00000100073
ENSDART00000141723 |
dyrk1aa
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A, a |
| chr13_+_31205439 | 0.39 |
ENSDART00000132326
|
ptpn20
|
protein tyrosine phosphatase, non-receptor type 20 |
| chr4_+_20177526 | 0.39 |
ENSDART00000017947
ENSDART00000135451 |
ccdc146
|
coiled-coil domain containing 146 |
| chr23_+_2760573 | 0.39 |
ENSDART00000129719
|
top1
|
DNA topoisomerase I |
| chr11_-_30352333 | 0.39 |
ENSDART00000030794
|
tmem169a
|
transmembrane protein 169a |
| chr13_+_36958086 | 0.39 |
ENSDART00000024386
|
frmd6
|
FERM domain containing 6 |
| chr2_-_58183499 | 0.38 |
ENSDART00000172281
ENSDART00000186262 |
si:ch1073-185p12.2
|
si:ch1073-185p12.2 |
| chr10_+_2807136 | 0.38 |
ENSDART00000126440
ENSDART00000131435 |
aebp1
|
AE binding protein 1 |
| chr9_-_40873934 | 0.37 |
ENSDART00000066424
|
pofut2
|
protein O-fucosyltransferase 2 |
| chr8_-_9570511 | 0.37 |
ENSDART00000044000
|
plxna3
|
plexin A3 |
| chr20_-_7582936 | 0.37 |
ENSDART00000083890
|
usp24
|
ubiquitin specific peptidase 24 |
| chr18_+_16192083 | 0.37 |
ENSDART00000133042
|
lrriq1
|
leucine-rich repeats and IQ motif containing 1 |
| chr6_-_23931442 | 0.37 |
ENSDART00000160547
|
sec16b
|
SEC16 homolog B, endoplasmic reticulum export factor |
| chr20_+_30797329 | 0.36 |
ENSDART00000145066
|
nhsl1b
|
NHS-like 1b |
| chr11_-_44194132 | 0.35 |
ENSDART00000182954
ENSDART00000111271 |
CABZ01080074.1
|
|
| chr4_+_77971104 | 0.35 |
ENSDART00000188609
|
zgc:113921
|
zgc:113921 |
| chr3_-_60175470 | 0.35 |
ENSDART00000156597
|
si:ch73-364h19.1
|
si:ch73-364h19.1 |
| chr5_+_25317061 | 0.35 |
ENSDART00000170097
|
trpm6
|
transient receptor potential cation channel, subfamily M, member 6 |
| chr10_-_44560165 | 0.35 |
ENSDART00000181217
ENSDART00000076084 |
npm2b
|
nucleophosmin/nucleoplasmin, 2b |
| chr16_-_31791165 | 0.34 |
ENSDART00000148389
|
chd4b
|
chromodomain helicase DNA binding protein 4b |
| chr7_+_23515966 | 0.34 |
ENSDART00000186893
ENSDART00000186189 |
zgc:109889
|
zgc:109889 |
| chr2_+_41526904 | 0.34 |
ENSDART00000127520
|
acvr1l
|
activin A receptor, type 1 like |
| chr8_+_25267903 | 0.34 |
ENSDART00000093090
|
ampd2b
|
adenosine monophosphate deaminase 2b |
| chr4_-_1801519 | 0.33 |
ENSDART00000188604
ENSDART00000135749 |
nudt4b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4b |
| chr2_+_33796207 | 0.33 |
ENSDART00000124647
|
kiss1ra
|
KISS1 receptor a |
| chr1_+_53279861 | 0.32 |
ENSDART00000035713
|
rnf150a
|
ring finger protein 150a |
| chr15_+_22267847 | 0.32 |
ENSDART00000110665
|
spa17
|
sperm autoantigenic protein 17 |
| chr14_+_10596628 | 0.32 |
ENSDART00000115177
|
gpr174
|
G protein-coupled receptor 174 |
| chr20_+_27003713 | 0.31 |
ENSDART00000153232
|
si:dkey-177p2.18
|
si:dkey-177p2.18 |
| chr5_+_61919217 | 0.31 |
ENSDART00000097340
|
rsph4a
|
radial spoke head 4 homolog A |
| chr5_-_72390259 | 0.31 |
ENSDART00000172302
|
wbp1
|
WW domain binding protein 1 |
| chr12_+_33361948 | 0.30 |
ENSDART00000124982
|
fasn
|
fatty acid synthase |
| chr14_+_7892021 | 0.30 |
ENSDART00000161307
|
ube2d2
|
ubiquitin-conjugating enzyme E2D 2 (UBC4/5 homolog, yeast) |
| chr13_-_14926318 | 0.30 |
ENSDART00000142785
|
cdc25b
|
cell division cycle 25B |
| chr22_-_16270462 | 0.30 |
ENSDART00000105681
|
cdc14ab
|
cell division cycle 14Ab |
| chr4_-_2545310 | 0.30 |
ENSDART00000150619
ENSDART00000140760 |
e2f7
|
E2F transcription factor 7 |
| chr7_-_21905851 | 0.30 |
ENSDART00000111066
ENSDART00000020288 |
epoa
|
erythropoietin a |
| chr6_-_58901552 | 0.29 |
ENSDART00000122003
|
mbd6
|
methyl-CpG binding domain protein 6 |
| chr25_-_21894706 | 0.29 |
ENSDART00000189158
|
fbxo31
|
F-box protein 31 |
| chr16_-_11779508 | 0.29 |
ENSDART00000136329
ENSDART00000060145 ENSDART00000141101 |
pafah1b3
|
platelet-activating factor acetylhydrolase, isoform Ib, gamma subunit |
| chr18_-_47103131 | 0.29 |
ENSDART00000188553
|
CABZ01039304.1
|
|
| chr22_-_16270071 | 0.29 |
ENSDART00000171823
|
cdc14ab
|
cell division cycle 14Ab |
| chr8_-_36412936 | 0.28 |
ENSDART00000159276
|
si:zfos-2070c2.3
|
si:zfos-2070c2.3 |
| chr19_+_2275019 | 0.28 |
ENSDART00000136138
|
itgb8
|
integrin, beta 8 |
| chr19_-_12324514 | 0.28 |
ENSDART00000122192
|
ncaldb
|
neurocalcin delta b |
| chr15_+_31735931 | 0.27 |
ENSDART00000185681
ENSDART00000149137 |
rxfp2b
|
relaxin/insulin-like family peptide receptor 2b |
| chr2_+_13462305 | 0.26 |
ENSDART00000149309
ENSDART00000080900 |
cfap57
|
cilia and flagella associated protein 57 |
| chr12_-_36521767 | 0.26 |
ENSDART00000110290
|
unc13d
|
unc-13 homolog D (C. elegans) |
| chr6_+_515181 | 0.25 |
ENSDART00000171374
|
CACNA1I (1 of many)
|
si:ch73-379f7.5 |
| chr20_-_28642061 | 0.25 |
ENSDART00000135513
|
rgs6
|
regulator of G protein signaling 6 |
| chr7_+_29163762 | 0.25 |
ENSDART00000173762
|
slc38a8b
|
solute carrier family 38, member 8b |
| chr4_+_51160367 | 0.24 |
ENSDART00000189984
|
znf1141
|
zinc finger protein 1141 |
| chr6_-_46560207 | 0.24 |
ENSDART00000187607
|
kcnb1
|
potassium voltage-gated channel, Shab-related subfamily, member 1 |
| chr7_-_29533127 | 0.24 |
ENSDART00000057289
|
anxa2b
|
annexin A2b |
| chr18_-_46763170 | 0.23 |
ENSDART00000171880
|
dner
|
delta/notch-like EGF repeat containing |
| chr16_+_13898399 | 0.23 |
ENSDART00000140946
|
si:ch211-149k23.9
|
si:ch211-149k23.9 |
| chr21_+_9576176 | 0.23 |
ENSDART00000161289
ENSDART00000159899 ENSDART00000162834 |
mapk10
|
mitogen-activated protein kinase 10 |
| chr20_-_38617766 | 0.23 |
ENSDART00000050474
|
slc30a2
|
solute carrier family 30 (zinc transporter), member 2 |
| chr21_-_435466 | 0.23 |
ENSDART00000110297
|
klf4
|
Kruppel-like factor 4 |
| chr2_+_59527149 | 0.22 |
ENSDART00000168568
|
CU861477.1
|
|
| chr24_+_39186940 | 0.22 |
ENSDART00000155817
|
spsb3b
|
splA/ryanodine receptor domain and SOCS box containing 3b |
| chr6_+_18251140 | 0.22 |
ENSDART00000169752
|
ccdc40
|
coiled-coil domain containing 40 |
| chr13_-_48756036 | 0.22 |
ENSDART00000035018
ENSDART00000132895 |
ntpcr
|
nucleoside-triphosphatase, cancer-related |
| chr13_+_12606821 | 0.22 |
ENSDART00000140096
ENSDART00000145136 |
metap1
|
methionyl aminopeptidase 1 |
| chr7_-_50395059 | 0.21 |
ENSDART00000191150
|
vps33b
|
vacuolar protein sorting 33B |
| chr19_-_27588842 | 0.21 |
ENSDART00000121643
|
si:dkeyp-46h3.2
|
si:dkeyp-46h3.2 |
| chr2_-_7696287 | 0.20 |
ENSDART00000190769
|
CABZ01055306.1
|
|
| chr13_-_48764180 | 0.20 |
ENSDART00000167157
|
si:ch1073-266p11.2
|
si:ch1073-266p11.2 |
| chr12_+_13405445 | 0.20 |
ENSDART00000089042
|
kcnh4b
|
potassium voltage-gated channel, subfamily H (eag-related), member 4b |
| chr3_+_60044780 | 0.20 |
ENSDART00000080437
|
zgc:113030
|
zgc:113030 |
| chr5_-_69940868 | 0.19 |
ENSDART00000185924
ENSDART00000097357 |
ugt2a4
|
UDP glucuronosyltransferase 2 family, polypeptide A4 |
| chr18_+_38191346 | 0.19 |
ENSDART00000052703
|
nucb2b
|
nucleobindin 2b |
| chr21_-_43606502 | 0.19 |
ENSDART00000151030
|
si:ch73-362m14.4
|
si:ch73-362m14.4 |
| chr17_-_8173380 | 0.18 |
ENSDART00000148520
|
syne1b
|
spectrin repeat containing, nuclear envelope 1b |
| chr20_+_27003558 | 0.18 |
ENSDART00000125688
|
si:dkey-177p2.18
|
si:dkey-177p2.18 |
| chr10_+_8101729 | 0.18 |
ENSDART00000138875
ENSDART00000123447 ENSDART00000185333 |
pstpip2
|
proline-serine-threonine phosphatase interacting protein 2 |
| chr2_-_10631767 | 0.18 |
ENSDART00000190033
|
mtf2
|
metal response element binding transcription factor 2 |
| chr4_-_12795436 | 0.18 |
ENSDART00000131026
ENSDART00000075127 |
b2m
|
beta-2-microglobulin |
| chr17_+_53186347 | 0.18 |
ENSDART00000155981
|
dph6
|
diphthamine biosynthesis 6 |
| chr7_-_19999152 | 0.18 |
ENSDART00000173881
ENSDART00000100798 |
trip6
|
thyroid hormone receptor interactor 6 |
| chr24_-_37003577 | 0.18 |
ENSDART00000137777
|
dnaaf3
|
dynein, axonemal, assembly factor 3 |
| chr19_-_12315693 | 0.17 |
ENSDART00000151158
|
ncaldb
|
neurocalcin delta b |
| chr20_+_4060839 | 0.17 |
ENSDART00000178565
|
TRIM67
|
tripartite motif containing 67 |
| chr25_-_6557854 | 0.17 |
ENSDART00000181740
|
cspg4
|
chondroitin sulfate proteoglycan 4 |
| chr24_-_2843107 | 0.17 |
ENSDART00000165290
|
cyb5a
|
cytochrome b5 type A (microsomal) |
| chr7_+_38445938 | 0.17 |
ENSDART00000173467
|
cep89
|
centrosomal protein 89 |
| chr11_+_77526 | 0.16 |
ENSDART00000193521
|
CABZ01072242.1
|
|
| chr22_-_37565348 | 0.16 |
ENSDART00000149482
ENSDART00000104478 |
fxr1
|
fragile X mental retardation, autosomal homolog 1 |
| chr7_+_27059330 | 0.16 |
ENSDART00000173919
|
plekha7b
|
pleckstrin homology domain containing, family A member 7b |
| chr19_+_2279051 | 0.16 |
ENSDART00000182103
|
itgb8
|
integrin, beta 8 |
| chr16_-_17525322 | 0.16 |
ENSDART00000189720
|
clcn1b
|
chloride channel, voltage-sensitive 1b |
| chr6_+_27361978 | 0.16 |
ENSDART00000065238
|
crocc2
|
ciliary rootlet coiled-coil, rootletin family member 2 |
| chr1_-_49498116 | 0.16 |
ENSDART00000137357
|
zgc:175214
|
zgc:175214 |
| chr13_+_9368621 | 0.15 |
ENSDART00000109126
|
alms1
|
Alstrom syndrome protein 1 |
| chr17_-_51651631 | 0.15 |
ENSDART00000154699
|
ccr6b
|
chemokine (C-C motif) receptor 6b |
| chr13_-_7767548 | 0.15 |
ENSDART00000158720
ENSDART00000121952 |
h2afy2
|
H2A histone family, member Y2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of rfx1a+rfx1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.3 | 0.9 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.2 | 0.7 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.2 | 0.7 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.2 | 0.9 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.2 | 0.8 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.2 | 1.0 | GO:0048855 | adenohypophysis morphogenesis(GO:0048855) |
| 0.2 | 0.5 | GO:0097053 | L-kynurenine metabolic process(GO:0097052) L-kynurenine catabolic process(GO:0097053) |
| 0.2 | 1.6 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.2 | 0.7 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.2 | 1.1 | GO:0098815 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.1 | 0.8 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 0.5 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.1 | 1.0 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.1 | 0.4 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.1 | 0.3 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.1 | 0.2 | GO:0014014 | negative regulation of gliogenesis(GO:0014014) |
| 0.1 | 0.6 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.1 | 0.8 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 0.4 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.6 | GO:0070293 | renal absorption(GO:0070293) |
| 0.1 | 0.9 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 0.9 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.1 | 0.3 | GO:1901909 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.5 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.4 | GO:0009092 | homoserine metabolic process(GO:0009092) cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) transsulfuration(GO:0019346) |
| 0.1 | 0.4 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 0.4 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.1 | 0.3 | GO:0045448 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.1 | 0.2 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.1 | 0.3 | GO:0032875 | regulation of DNA endoreduplication(GO:0032875) positive regulation of DNA endoreduplication(GO:0032877) DNA endoreduplication(GO:0042023) |
| 0.1 | 0.6 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.1 | 0.2 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.1 | 1.1 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.0 | 0.9 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.2 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.3 | GO:0048823 | nucleate erythrocyte development(GO:0048823) |
| 0.0 | 0.6 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.4 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.5 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.3 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.0 | 0.9 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.5 | GO:0050482 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.9 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.5 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.2 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 0.2 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.1 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 | 0.5 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 1.0 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.0 | 0.3 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 0.3 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.4 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.1 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.7 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.2 | GO:1901224 | positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.0 | 0.1 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.0 | 0.3 | GO:0030819 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.0 | 0.6 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.2 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.1 | GO:0043393 | regulation of protein binding(GO:0043393) |
| 0.0 | 0.1 | GO:0090177 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 1.7 | GO:0031098 | stress-activated protein kinase signaling cascade(GO:0031098) |
| 0.0 | 0.2 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 0.0 | 0.4 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 0.2 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:0072091 | regulation of stem cell proliferation(GO:0072091) |
| 0.0 | 0.4 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 0.9 | GO:1990542 | mitochondrial transmembrane transport(GO:1990542) |
| 0.0 | 0.4 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.1 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.2 | 1.0 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 0.3 | GO:0001534 | radial spoke(GO:0001534) |
| 0.1 | 0.3 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 0.4 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 1.1 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 0.3 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.1 | 0.5 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 0.6 | GO:0099634 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.2 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.2 | GO:1902737 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.0 | 0.9 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 1.3 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.8 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.4 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.9 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.1 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 1.6 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 1.6 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 3.3 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 1.1 | GO:0008328 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 0.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.2 | 0.7 | GO:0071253 | connexin binding(GO:0071253) |
| 0.2 | 0.5 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.2 | 0.8 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.2 | 1.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.9 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 0.7 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 0.5 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.1 | 0.9 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.1 | 0.7 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.1 | 0.3 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.1 | 0.7 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 0.3 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 0.7 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.5 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 0.6 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.9 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 0.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.6 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 0.3 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.0 | 0.5 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.8 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.7 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.3 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.4 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.4 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.5 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.3 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.9 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 3.9 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.4 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.2 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.5 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.2 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.0 | 0.9 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 1.9 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.4 | GO:1901682 | sulfur compound transmembrane transporter activity(GO:1901682) |
| 0.0 | 0.8 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 0.2 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 1.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.7 | GO:0042562 | hormone binding(GO:0042562) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.8 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.2 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.1 | 0.4 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.4 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.7 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.2 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.5 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.3 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.8 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.3 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.8 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.2 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.5 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |