Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
Results for rel
Z-value: 0.56
Transcription factors associated with rel
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rel
|
ENSDARG00000055276 | v-rel avian reticuloendotheliosis viral oncogene homolog |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rel | dr11_v1_chr13_-_25819825_25819825 | -0.06 | 8.1e-01 | Click! |
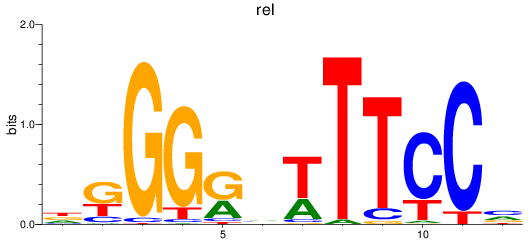
Activity profile of rel motif
Sorted Z-values of rel motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_22843562 | 1.05 |
ENSDART00000142738
|
pbld
|
phenazine biosynthesis like protein domain containing |
| chr8_+_47633438 | 1.04 |
ENSDART00000139096
|
si:ch211-251b21.1
|
si:ch211-251b21.1 |
| chr1_+_12135129 | 1.02 |
ENSDART00000126020
|
spink4
|
serine peptidase inhibitor, Kazal type 4 |
| chr2_-_51772438 | 1.02 |
ENSDART00000170241
|
BX908782.2
|
Danio rerio three-finger protein 5 (LOC100003647), mRNA. |
| chr2_+_45191049 | 0.92 |
ENSDART00000165392
|
ccl20a.3
|
chemokine (C-C motif) ligand 20a, duplicate 3 |
| chr20_+_33981946 | 0.87 |
ENSDART00000131775
|
si:dkey-51e6.1
|
si:dkey-51e6.1 |
| chr11_-_3552067 | 0.84 |
ENSDART00000163656
|
CAMK2N1
|
si:dkey-33m11.6 |
| chr2_+_42191592 | 0.79 |
ENSDART00000144716
|
cavin4a
|
caveolae associated protein 4a |
| chr17_+_20205185 | 0.77 |
ENSDART00000167007
|
gnrh3
|
gonadotropin-releasing hormone 3 |
| chr14_+_36885524 | 0.77 |
ENSDART00000032547
|
lect2l
|
leukocyte cell-derived chemotaxin 2 like |
| chr2_-_28671139 | 0.76 |
ENSDART00000165272
ENSDART00000164657 |
dhcr7
|
7-dehydrocholesterol reductase |
| chr3_-_21242460 | 0.74 |
ENSDART00000007293
|
tcap
|
titin-cap (telethonin) |
| chr8_+_48603398 | 0.73 |
ENSDART00000074900
|
zgc:195023
|
zgc:195023 |
| chr10_-_39011514 | 0.72 |
ENSDART00000075123
|
pcp4a
|
Purkinje cell protein 4a |
| chr24_+_26006730 | 0.69 |
ENSDART00000140384
ENSDART00000139184 |
ccl20b
|
chemokine (C-C motif) ligand 20b |
| chr20_+_16881883 | 0.64 |
ENSDART00000130107
|
nfkbiaa
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha a |
| chr1_+_41690402 | 0.64 |
ENSDART00000177298
|
fbxo41
|
F-box protein 41 |
| chr8_-_14049404 | 0.62 |
ENSDART00000093117
|
atp2b3a
|
ATPase plasma membrane Ca2+ transporting 3a |
| chr7_-_20241346 | 0.61 |
ENSDART00000173619
ENSDART00000127699 |
si:ch73-335l21.4
|
si:ch73-335l21.4 |
| chr1_+_33969015 | 0.60 |
ENSDART00000042984
ENSDART00000146530 |
epha6
|
eph receptor A6 |
| chr17_+_8175998 | 0.60 |
ENSDART00000131200
|
myct1b
|
myc target 1b |
| chr11_+_25634041 | 0.59 |
ENSDART00000033657
|
grm6b
|
glutamate receptor, metabotropic 6b |
| chr7_+_45975537 | 0.58 |
ENSDART00000170253
|
plekhf1
|
pleckstrin homology domain containing, family F (with FYVE domain) member 1 |
| chr21_-_2348838 | 0.58 |
ENSDART00000160337
|
si:ch73-299h12.8
|
si:ch73-299h12.8 |
| chr13_+_27316934 | 0.57 |
ENSDART00000164533
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr14_+_20893065 | 0.57 |
ENSDART00000079452
|
lygl1
|
lysozyme g-like 1 |
| chr22_+_5478353 | 0.54 |
ENSDART00000160596
|
tppp
|
tubulin polymerization promoting protein |
| chr3_+_46459540 | 0.53 |
ENSDART00000188150
|
si:ch211-66e2.5
|
si:ch211-66e2.5 |
| chr16_-_12984631 | 0.53 |
ENSDART00000184863
|
cacng7b
|
calcium channel, voltage-dependent, gamma subunit 7b |
| chr14_-_11529311 | 0.53 |
ENSDART00000127208
|
si:ch211-153b23.7
|
si:ch211-153b23.7 |
| chr2_+_2737422 | 0.52 |
ENSDART00000032459
|
aqp1a.1
|
aquaporin 1a (Colton blood group), tandem duplicate 1 |
| chr4_-_18954001 | 0.52 |
ENSDART00000144814
|
si:dkey-31f5.8
|
si:dkey-31f5.8 |
| chr10_+_33744098 | 0.52 |
ENSDART00000147775
|
rxfp2a
|
relaxin/insulin-like family peptide receptor 2a |
| chr2_+_6963296 | 0.51 |
ENSDART00000147146
|
ddr2b
|
discoidin domain receptor tyrosine kinase 2b |
| chr25_+_37397031 | 0.51 |
ENSDART00000193643
ENSDART00000169132 |
slc1a2b
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2b |
| chr25_-_35960229 | 0.50 |
ENSDART00000073434
|
snx20
|
sorting nexin 20 |
| chr14_+_3038473 | 0.50 |
ENSDART00000026021
ENSDART00000150000 |
cd74a
|
CD74 molecule, major histocompatibility complex, class II invariant chain a |
| chr12_-_3903886 | 0.49 |
ENSDART00000184214
ENSDART00000041082 |
gdpd3b
|
glycerophosphodiester phosphodiesterase domain containing 3b |
| chr24_-_12689571 | 0.49 |
ENSDART00000015517
|
pdcd6
|
programmed cell death 6 |
| chr17_+_12942634 | 0.49 |
ENSDART00000016597
|
nfkbiab
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha b |
| chr16_+_32995882 | 0.49 |
ENSDART00000170157
|
prss35
|
protease, serine, 35 |
| chr13_+_27316632 | 0.48 |
ENSDART00000016121
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr14_+_36889893 | 0.45 |
ENSDART00000124159
|
si:ch211-132p1.3
|
si:ch211-132p1.3 |
| chr5_-_30984271 | 0.44 |
ENSDART00000051392
|
spns3
|
spinster homolog 3 (Drosophila) |
| chr1_+_16396870 | 0.44 |
ENSDART00000189245
ENSDART00000113266 |
micu3a
|
mitochondrial calcium uptake family, member 3a |
| chr11_-_7156620 | 0.44 |
ENSDART00000172823
ENSDART00000172879 ENSDART00000078916 |
smim7
|
small integral membrane protein 7 |
| chr3_+_11568523 | 0.44 |
ENSDART00000179668
|
CABZ01087224.1
|
|
| chr20_+_36234335 | 0.44 |
ENSDART00000193484
ENSDART00000181664 |
cnih3
|
cornichon family AMPA receptor auxiliary protein 3 |
| chr4_+_18806251 | 0.43 |
ENSDART00000138662
|
slc26a3.2
|
solute carrier family 26 (anion exchanger), member 3, tandem duplicate 2 |
| chr2_-_29485408 | 0.43 |
ENSDART00000013411
|
cahz
|
carbonic anhydrase |
| chr4_-_33525219 | 0.43 |
ENSDART00000150491
|
CT027801.2
|
|
| chr25_-_13188214 | 0.42 |
ENSDART00000187298
|
si:ch211-147m6.1
|
si:ch211-147m6.1 |
| chr20_+_22045089 | 0.41 |
ENSDART00000063564
ENSDART00000187013 ENSDART00000161552 ENSDART00000174478 ENSDART00000063568 ENSDART00000152247 |
nmu
|
neuromedin U |
| chr18_-_17077419 | 0.41 |
ENSDART00000148714
|
il17c
|
interleukin 17c |
| chr22_+_110158 | 0.41 |
ENSDART00000143698
|
prkar2ab
|
protein kinase, cAMP-dependent, regulatory, type II, alpha, B |
| chr20_-_44496245 | 0.40 |
ENSDART00000012229
|
fkbp1b
|
FK506 binding protein 1b |
| chr1_+_16397063 | 0.40 |
ENSDART00000159794
|
micu3a
|
mitochondrial calcium uptake family, member 3a |
| chr25_-_27843066 | 0.39 |
ENSDART00000179684
ENSDART00000186000 ENSDART00000190065 |
asb15a
|
ankyrin repeat and SOCS box containing 15a |
| chr8_-_39749407 | 0.39 |
ENSDART00000180336
|
CT027598.1
|
|
| chr3_-_52644737 | 0.39 |
ENSDART00000126180
|
si:dkey-210j14.3
|
si:dkey-210j14.3 |
| chr24_-_29997145 | 0.38 |
ENSDART00000135094
|
palmdb
|
palmdelphin b |
| chr8_+_7778770 | 0.38 |
ENSDART00000171325
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr3_+_34670076 | 0.37 |
ENSDART00000133457
|
dlx4a
|
distal-less homeobox 4a |
| chr11_-_19694334 | 0.36 |
ENSDART00000054735
|
SYNPR
|
si:dkey-30j16.3 |
| chr15_+_37436430 | 0.35 |
ENSDART00000124779
|
igflr1
|
IGF-like family receptor 1 |
| chr10_+_26926654 | 0.35 |
ENSDART00000078980
ENSDART00000100289 |
rab1bb
|
RAB1B, member RAS oncogene family b |
| chr12_+_7445595 | 0.35 |
ENSDART00000103536
ENSDART00000152524 |
phyhiplb
|
phytanoyl-CoA 2-hydroxylase interacting protein-like b |
| chr15_+_42235449 | 0.33 |
ENSDART00000114801
ENSDART00000182053 |
SGPP2
|
sphingosine-1-phosphate phosphatase 2 |
| chr2_-_9857932 | 0.33 |
ENSDART00000154490
|
si:ch211-252f13.5
|
si:ch211-252f13.5 |
| chr13_-_2010191 | 0.33 |
ENSDART00000161021
ENSDART00000124134 |
gfral
|
GDNF family receptor alpha like |
| chr2_-_32501501 | 0.33 |
ENSDART00000181309
|
faim2a
|
Fas apoptotic inhibitory molecule 2a |
| chr19_-_7043355 | 0.32 |
ENSDART00000104845
|
tapbp.1
|
TAP binding protein (tapasin), tandem duplicate 1 |
| chr14_-_30967284 | 0.32 |
ENSDART00000149435
|
il2rgb
|
interleukin 2 receptor, gamma b |
| chr11_+_40544269 | 0.32 |
ENSDART00000175424
ENSDART00000184673 |
mmel1
|
membrane metallo-endopeptidase-like 1 |
| chr22_+_14051245 | 0.32 |
ENSDART00000043711
ENSDART00000164259 |
aox6
|
aldehyde oxidase 6 |
| chr25_+_8955530 | 0.31 |
ENSDART00000156444
|
si:ch211-256a21.4
|
si:ch211-256a21.4 |
| chr8_-_46572298 | 0.31 |
ENSDART00000030470
|
sult1st3
|
sulfotransferase family 1, cytosolic sulfotransferase 3 |
| chr14_+_26796917 | 0.31 |
ENSDART00000137695
|
klhl4
|
kelch-like family member 4 |
| chr1_-_49225890 | 0.31 |
ENSDART00000111598
|
cxcl18b
|
chemokine (C-X-C motif) ligand 18b |
| chr9_-_23894392 | 0.31 |
ENSDART00000133417
|
asb18
|
ankyrin repeat and SOCS box containing 18 |
| chr24_-_18477672 | 0.30 |
ENSDART00000126928
ENSDART00000158393 |
si:dkey-73n8.3
|
si:dkey-73n8.3 |
| chr23_-_24190712 | 0.29 |
ENSDART00000139514
|
ano11
|
anoctamin 11 |
| chr16_-_41004731 | 0.29 |
ENSDART00000102591
|
KCNV1
|
si:dkey-201i6.2 |
| chr9_-_5263947 | 0.29 |
ENSDART00000088342
|
cytip
|
cytohesin 1 interacting protein |
| chr23_-_35069805 | 0.28 |
ENSDART00000087219
|
BX294434.1
|
|
| chr24_-_18467064 | 0.28 |
ENSDART00000081946
|
zgc:112332
|
zgc:112332 |
| chr1_-_49409549 | 0.28 |
ENSDART00000074317
|
GSK3B (1 of many)
|
si:dkeyp-80c12.7 |
| chr22_+_661711 | 0.28 |
ENSDART00000113795
|
elf3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr25_-_28926631 | 0.28 |
ENSDART00000112850
|
etfbkmt
|
electron transfer flavoprotein beta subunit lysine methyltransferase |
| chr20_+_28434196 | 0.28 |
ENSDART00000034245
|
dpf3
|
D4, zinc and double PHD fingers, family 3 |
| chr1_+_51896147 | 0.27 |
ENSDART00000167139
|
nfixa
|
nuclear factor I/Xa |
| chr6_-_54815886 | 0.27 |
ENSDART00000180793
ENSDART00000007498 |
tnni1b
|
troponin I type 1b (skeletal, slow) |
| chr19_-_42249597 | 0.27 |
ENSDART00000184762
|
CR936363.1
|
|
| chr4_-_25485404 | 0.27 |
ENSDART00000041402
|
pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr23_+_3620618 | 0.27 |
ENSDART00000190126
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr2_-_53592532 | 0.26 |
ENSDART00000184066
|
ccl25a
|
chemokine (C-C motif) ligand 25a |
| chr5_+_13385837 | 0.25 |
ENSDART00000191190
|
ccl19a.1
|
chemokine (C-C motif) ligand 19a, tandem duplicate 1 |
| chr22_+_6293563 | 0.25 |
ENSDART00000063416
|
rnasel2
|
ribonuclease like 2 |
| chr9_+_31222026 | 0.25 |
ENSDART00000145573
|
clybl
|
citrate lyase beta like |
| chr18_-_49286381 | 0.24 |
ENSDART00000174248
ENSDART00000174038 |
si:zfos-464b6.2
|
si:zfos-464b6.2 |
| chr4_-_25485214 | 0.24 |
ENSDART00000159941
|
pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr24_-_33703504 | 0.24 |
ENSDART00000079292
|
cavin4b
|
caveolae associated protein 4b |
| chr6_+_3334710 | 0.24 |
ENSDART00000132848
|
st3gal3a
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3a |
| chr7_+_39634873 | 0.24 |
ENSDART00000114774
|
ptpn5
|
protein tyrosine phosphatase, non-receptor type 5 |
| chr23_+_3618454 | 0.24 |
ENSDART00000189393
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr20_-_8321280 | 0.24 |
ENSDART00000083906
ENSDART00000186477 |
dab1a
|
Dab, reelin signal transducer, homolog 1a (Drosophila) |
| chr6_+_3334392 | 0.23 |
ENSDART00000133707
ENSDART00000130879 |
st3gal3a
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3a |
| chr10_-_14536399 | 0.23 |
ENSDART00000186501
|
hdhd2
|
haloacid dehalogenase-like hydrolase domain containing 2 |
| chr14_+_6182346 | 0.23 |
ENSDART00000075525
|
si:ch73-22a13.3
|
si:ch73-22a13.3 |
| chr23_+_3591690 | 0.23 |
ENSDART00000180822
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr3_+_54744069 | 0.22 |
ENSDART00000134958
ENSDART00000114443 |
si:ch211-74m13.3
|
si:ch211-74m13.3 |
| chr7_+_39706004 | 0.22 |
ENSDART00000161856
|
ccl36.1
|
chemokine (C-C motif) ligand 36, duplicate 1 |
| chr23_+_23183449 | 0.22 |
ENSDART00000132296
|
klhl17
|
kelch-like family member 17 |
| chr23_+_3596401 | 0.22 |
ENSDART00000185908
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr13_+_41819817 | 0.22 |
ENSDART00000185778
|
CABZ01066611.1
|
|
| chr22_+_2207502 | 0.22 |
ENSDART00000169162
|
si:dkeyp-79b7.12
|
si:dkeyp-79b7.12 |
| chr1_+_10771457 | 0.21 |
ENSDART00000027050
ENSDART00000137185 |
cnga3b
|
cyclic nucleotide gated channel alpha 3b |
| chr4_-_12790886 | 0.21 |
ENSDART00000182535
ENSDART00000067131 ENSDART00000186426 |
irak3
|
interleukin-1 receptor-associated kinase 3 |
| chr8_+_24281512 | 0.21 |
ENSDART00000062845
|
mmp9
|
matrix metallopeptidase 9 |
| chr10_+_20630444 | 0.21 |
ENSDART00000191182
|
prnpb
|
prion protein b |
| chr7_+_53154946 | 0.20 |
ENSDART00000185104
ENSDART00000191559 ENSDART00000160096 |
cdh29
|
cadherin 29 |
| chr17_+_24613255 | 0.20 |
ENSDART00000064738
|
atp5if1b
|
ATP synthase inhibitory factor subunit 1b |
| chr16_+_23403602 | 0.20 |
ENSDART00000159848
|
s100w
|
S100 calcium binding protein W |
| chr23_+_3594171 | 0.19 |
ENSDART00000159609
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr13_-_27916439 | 0.19 |
ENSDART00000139081
ENSDART00000087097 |
ogfrl1
|
opioid growth factor receptor-like 1 |
| chr5_+_24882633 | 0.19 |
ENSDART00000111302
|
rhbdd3
|
rhomboid domain containing 3 |
| chr23_+_19701587 | 0.19 |
ENSDART00000104425
|
dnase1l1
|
deoxyribonuclease I-like 1 |
| chr13_-_40120252 | 0.19 |
ENSDART00000157852
|
crtac1b
|
cartilage acidic protein 1b |
| chr23_-_36418059 | 0.19 |
ENSDART00000135232
|
znf740b
|
zinc finger protein 740b |
| chr18_-_5527050 | 0.18 |
ENSDART00000145400
ENSDART00000132498 ENSDART00000146209 |
zgc:153317
|
zgc:153317 |
| chr18_-_18543358 | 0.18 |
ENSDART00000126460
|
il34
|
interleukin 34 |
| chr7_-_57905274 | 0.18 |
ENSDART00000145925
|
ank2b
|
ankyrin 2b, neuronal |
| chr12_-_30760971 | 0.18 |
ENSDART00000066257
|
entpd1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr22_+_8536838 | 0.18 |
ENSDART00000132998
|
si:ch73-27e22.7
|
si:ch73-27e22.7 |
| chr18_-_17077596 | 0.17 |
ENSDART00000111821
|
il17c
|
interleukin 17c |
| chr14_+_30773764 | 0.17 |
ENSDART00000186961
|
atl3
|
atlastin 3 |
| chr15_+_6459847 | 0.17 |
ENSDART00000157250
ENSDART00000065824 |
bace2
|
beta-site APP-cleaving enzyme 2 |
| chr1_-_52447364 | 0.17 |
ENSDART00000140740
|
si:ch211-217k17.10
|
si:ch211-217k17.10 |
| chr8_-_18667693 | 0.17 |
ENSDART00000100516
|
stap2b
|
signal transducing adaptor family member 2b |
| chr22_+_3045495 | 0.16 |
ENSDART00000164061
|
LO017843.1
|
|
| chr6_+_2195625 | 0.16 |
ENSDART00000155659
|
acvr1bb
|
activin A receptor type 1Bb |
| chr6_-_10305918 | 0.16 |
ENSDART00000090994
|
scn1lab
|
sodium channel, voltage-gated, type I like, alpha b |
| chr23_-_29502287 | 0.16 |
ENSDART00000141075
ENSDART00000053807 |
kif1b
|
kinesin family member 1B |
| chr13_-_45795722 | 0.16 |
ENSDART00000141779
|
fndc5a
|
fibronectin type III domain containing 5a |
| chr17_+_27803608 | 0.16 |
ENSDART00000164943
|
qkia
|
QKI, KH domain containing, RNA binding a |
| chr18_+_19883325 | 0.16 |
ENSDART00000182625
|
zgc:162898
|
zgc:162898 |
| chr11_-_40101246 | 0.15 |
ENSDART00000161083
|
tnfrsf9b
|
tumor necrosis factor receptor superfamily, member 9b |
| chr17_+_27158706 | 0.15 |
ENSDART00000151829
|
rps6ka1
|
ribosomal protein S6 kinase a, polypeptide 1 |
| chr13_+_2538636 | 0.15 |
ENSDART00000168927
|
plpp4
|
phospholipid phosphatase 4 |
| chr4_-_149334 | 0.15 |
ENSDART00000163280
|
tbk1
|
TANK-binding kinase 1 |
| chr5_+_58996765 | 0.15 |
ENSDART00000062175
|
derl2
|
derlin 2 |
| chr2_+_30878864 | 0.15 |
ENSDART00000009326
|
oprk1
|
opioid receptor, kappa 1 |
| chr4_-_12795436 | 0.14 |
ENSDART00000131026
ENSDART00000075127 |
b2m
|
beta-2-microglobulin |
| chr16_+_38394371 | 0.14 |
ENSDART00000137954
|
cd83
|
CD83 molecule |
| chr16_+_26846495 | 0.14 |
ENSDART00000078124
|
trim35-29
|
tripartite motif containing 35-29 |
| chr24_+_39137001 | 0.14 |
ENSDART00000181086
ENSDART00000183724 ENSDART00000193466 |
tbc1d24
|
TBC1 domain family, member 24 |
| chr12_-_19250854 | 0.13 |
ENSDART00000152844
|
cdc42ep1a
|
CDC42 effector protein (Rho GTPase binding) 1a |
| chr7_+_29293452 | 0.13 |
ENSDART00000127358
|
si:ch211-112g6.4
|
si:ch211-112g6.4 |
| chr7_-_73843720 | 0.12 |
ENSDART00000111622
|
caap1
|
caspase activity and apoptosis inhibitor 1 |
| chr11_+_6456146 | 0.12 |
ENSDART00000036939
|
gadd45ba
|
growth arrest and DNA-damage-inducible, beta a |
| chr8_+_39674707 | 0.12 |
ENSDART00000126301
ENSDART00000040330 |
prkab1b
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit, b |
| chr16_+_13855039 | 0.12 |
ENSDART00000113764
ENSDART00000143983 |
zgc:174888
|
zgc:174888 |
| chr16_+_22864722 | 0.12 |
ENSDART00000137783
|
flad1
|
flavin adenine dinucleotide synthetase 1 |
| chr22_+_26263290 | 0.12 |
ENSDART00000184840
|
BX649315.1
|
|
| chr15_-_37779978 | 0.12 |
ENSDART00000157202
|
si:dkey-42l23.3
|
si:dkey-42l23.3 |
| chr11_-_29737088 | 0.12 |
ENSDART00000159828
|
si:ch211-218g23.6
|
si:ch211-218g23.6 |
| chr7_-_19364813 | 0.12 |
ENSDART00000173977
|
ntn4
|
netrin 4 |
| chr17_+_48164536 | 0.11 |
ENSDART00000161750
ENSDART00000156923 |
plekhd1
|
pleckstrin homology domain containing, family D (with coiled-coil domains) member 1 |
| chr9_+_54695981 | 0.11 |
ENSDART00000183605
|
rab9a
|
RAB9A, member RAS oncogene family |
| chr18_+_5514269 | 0.11 |
ENSDART00000182614
|
slc30a4
|
solute carrier family 30 (zinc transporter), member 4 |
| chr13_+_21601149 | 0.11 |
ENSDART00000179369
|
sh2d4ba
|
SH2 domain containing 4Ba |
| chr1_-_7917062 | 0.11 |
ENSDART00000177068
|
mmd2b
|
monocyte to macrophage differentiation-associated 2b |
| chr12_+_18458502 | 0.11 |
ENSDART00000108745
|
rnf151
|
ring finger protein 151 |
| chr16_-_26296477 | 0.11 |
ENSDART00000157553
|
erfl1
|
Ets2 repressor factor like 1 |
| chr5_+_45138934 | 0.10 |
ENSDART00000041412
ENSDART00000136002 |
smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr7_+_56472585 | 0.10 |
ENSDART00000135259
ENSDART00000073596 |
ist1
|
increased sodium tolerance 1 homolog (yeast) |
| chr23_-_18707418 | 0.10 |
ENSDART00000144668
ENSDART00000141205 ENSDART00000016765 |
zgc:103759
|
zgc:103759 |
| chr13_+_31182103 | 0.10 |
ENSDART00000137664
|
ptpn20
|
protein tyrosine phosphatase, non-receptor type 20 |
| chr1_+_27690 | 0.10 |
ENSDART00000162928
|
eed
|
embryonic ectoderm development |
| chr21_+_21679086 | 0.10 |
ENSDART00000146225
|
or125-5
|
odorant receptor, family E, subfamily 125, member 5 |
| chr16_-_22225295 | 0.10 |
ENSDART00000163519
|
LO017682.1
|
|
| chr18_+_7073130 | 0.10 |
ENSDART00000101216
ENSDART00000148947 |
si:dkey-88e18.2
|
si:dkey-88e18.2 |
| chr8_+_52642869 | 0.10 |
ENSDART00000163617
ENSDART00000189997 |
si:dkey-90l8.3
|
si:dkey-90l8.3 |
| chr14_+_16765992 | 0.10 |
ENSDART00000140061
|
sqstm1
|
sequestosome 1 |
| chr20_-_29864390 | 0.10 |
ENSDART00000161834
ENSDART00000132278 |
rnf144ab
|
ring finger protein 144ab |
| chr22_+_25733817 | 0.10 |
ENSDART00000176962
ENSDART00000187865 |
si:dkeyp-98a7.9
|
si:dkeyp-98a7.9 |
| chr22_-_23067859 | 0.09 |
ENSDART00000137873
|
ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr3_-_31619463 | 0.09 |
ENSDART00000124559
|
moto
|
minamoto |
| chr2_+_42344304 | 0.09 |
ENSDART00000136105
|
ftr10
|
finTRIM family, member 10 |
| chr25_-_28926330 | 0.09 |
ENSDART00000155173
|
etfbkmt
|
electron transfer flavoprotein beta subunit lysine methyltransferase |
| chr6_-_442163 | 0.09 |
ENSDART00000163564
ENSDART00000189134 ENSDART00000169789 |
grap2b
|
GRB2-related adaptor protein 2b |
| chr2_-_50966124 | 0.09 |
ENSDART00000170470
|
ghrhra
|
growth hormone releasing hormone receptor a |
| chr25_-_7723685 | 0.09 |
ENSDART00000146882
|
phf21ab
|
PHD finger protein 21Ab |
| chr8_-_10949847 | 0.09 |
ENSDART00000123209
|
pqlc2
|
PQ loop repeat containing 2 |
| chr18_+_30847237 | 0.08 |
ENSDART00000012374
|
foxf1
|
forkhead box F1 |
| chr1_-_37377509 | 0.08 |
ENSDART00000113542
|
tnip2
|
TNFAIP3 interacting protein 2 |
| chr5_+_25596234 | 0.08 |
ENSDART00000143949
ENSDART00000167193 |
si:dkey-96l17.6
|
si:dkey-96l17.6 |
| chr22_-_651719 | 0.07 |
ENSDART00000148692
|
apobec2a
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2a |
| chr21_+_21817975 | 0.07 |
ENSDART00000151292
ENSDART00000166704 |
neu3.5
|
sialidase 3 (membrane sialidase), tandem duplicate 5 |
| chr13_-_133520 | 0.07 |
ENSDART00000180874
|
FO904997.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of rel
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0035971 | peptidyl-histidine dephosphorylation(GO:0035971) |
| 0.2 | 0.9 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.2 | 0.6 | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 0.2 | 0.6 | GO:0002468 | dendritic cell antigen processing and presentation(GO:0002468) |
| 0.2 | 0.6 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.2 | 0.7 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.1 | 0.4 | GO:0050995 | negative regulation of lipid catabolic process(GO:0050995) |
| 0.1 | 0.3 | GO:0072679 | thymocyte migration(GO:0072679) |
| 0.1 | 0.2 | GO:0071706 | tumor necrosis factor production(GO:0032640) tumor necrosis factor superfamily cytokine production(GO:0071706) |
| 0.1 | 0.2 | GO:0002631 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.1 | 0.2 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 0.2 | GO:0045649 | regulation of macrophage differentiation(GO:0045649) |
| 0.1 | 0.2 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 0.2 | GO:0048917 | posterior lateral line ganglion development(GO:0048917) |
| 0.0 | 0.6 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 1.9 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.8 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.5 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.0 | 0.1 | GO:0046443 | FAD biosynthetic process(GO:0006747) FAD metabolic process(GO:0046443) flavin adenine dinucleotide metabolic process(GO:0072387) flavin adenine dinucleotide biosynthetic process(GO:0072388) |
| 0.0 | 0.1 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.2 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.2 | GO:0033292 | T-tubule organization(GO:0033292) |
| 0.0 | 0.1 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.5 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.4 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.0 | 0.5 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.0 | 0.8 | GO:0042446 | hormone biosynthetic process(GO:0042446) |
| 0.0 | 0.2 | GO:0033986 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
| 0.0 | 0.2 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.3 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.0 | 0.4 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.1 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) |
| 0.0 | 0.2 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.4 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.9 | GO:0071222 | cellular response to molecule of bacterial origin(GO:0071219) cellular response to lipopolysaccharide(GO:0071222) |
| 0.0 | 0.1 | GO:0043388 | cytidine to uridine editing(GO:0016554) positive regulation of DNA binding(GO:0043388) |
| 0.0 | 0.1 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.2 | GO:0019883 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.4 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.5 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.2 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.5 | GO:0008345 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.0 | 0.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.3 | GO:0009712 | catecholamine metabolic process(GO:0006584) catechol-containing compound metabolic process(GO:0009712) |
| 0.0 | 1.0 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.2 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 0.1 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.1 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 0.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.2 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.0 | 1.0 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.0 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.2 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 0.8 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 0.5 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 1.0 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.1 | GO:0042611 | MHC protein complex(GO:0042611) MHC class I protein complex(GO:0042612) |
| 0.0 | 0.1 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.3 | GO:0071565 | nBAF complex(GO:0071565) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.3 | 0.8 | GO:0031530 | gonadotropin hormone-releasing hormone activity(GO:0005183) gonadotropin-releasing hormone receptor binding(GO:0031530) |
| 0.2 | 0.9 | GO:0101006 | protein histidine phosphatase activity(GO:0101006) |
| 0.2 | 0.6 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.1 | 0.6 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 0.3 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.1 | 0.6 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.1 | 0.3 | GO:0047777 | (3S)-citramalyl-CoA lyase activity(GO:0047777) |
| 0.1 | 0.5 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.1 | 0.5 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.1 | 0.4 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 0.5 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.1 | 2.2 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.1 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.1 | GO:0003919 | FMN adenylyltransferase activity(GO:0003919) |
| 0.0 | 0.3 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.5 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.6 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.3 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:1990174 | phosphodiesterase decapping endonuclease activity(GO:1990174) |
| 0.0 | 0.8 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.2 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.7 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.5 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.5 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 1.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.4 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.2 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.2 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.0 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.2 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.1 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.2 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.0 | 0.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |