Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
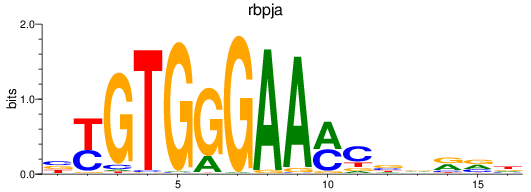
Results for rbpja
Z-value: 1.10
Transcription factors associated with rbpja
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rbpja
|
ENSDARG00000003398 | recombination signal binding protein for immunoglobulin kappa J region a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rbpja | dr11_v1_chr1_+_14454663_14454663 | 0.92 | 3.0e-08 | Click! |
Activity profile of rbpja motif
Sorted Z-values of rbpja motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_-_45705525 | 4.60 |
ENSDART00000148959
|
ednrab
|
endothelin receptor type Ab |
| chr3_+_41917499 | 4.05 |
ENSDART00000028673
|
lfng
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr6_+_30430591 | 3.70 |
ENSDART00000108943
|
shroom2a
|
shroom family member 2a |
| chr10_-_10864331 | 3.07 |
ENSDART00000122657
|
nrarpa
|
NOTCH regulated ankyrin repeat protein a |
| chr13_-_49819027 | 2.99 |
ENSDART00000067824
|
b3galnt2
|
beta-1,3-N-acetylgalactosaminyltransferase 2 |
| chr23_-_21446985 | 2.71 |
ENSDART00000044080
|
her12
|
hairy-related 12 |
| chr6_-_9695294 | 2.63 |
ENSDART00000162728
|
nop58
|
NOP58 ribonucleoprotein homolog (yeast) |
| chr10_-_10863936 | 2.61 |
ENSDART00000180568
|
nrarpa
|
NOTCH regulated ankyrin repeat protein a |
| chr1_-_23370395 | 2.51 |
ENSDART00000143014
ENSDART00000126785 ENSDART00000159138 |
pds5a
|
PDS5 cohesin associated factor A |
| chr19_+_42609132 | 2.47 |
ENSDART00000010104
|
crtap
|
cartilage associated protein |
| chr3_-_25119839 | 2.42 |
ENSDART00000154724
|
chadla
|
chondroadherin-like a |
| chr3_+_36646054 | 2.42 |
ENSDART00000170013
ENSDART00000159948 |
gspt1l
|
G1 to S phase transition 1, like |
| chr16_-_25680666 | 2.41 |
ENSDART00000132693
ENSDART00000140539 ENSDART00000015302 |
tomm40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr4_+_21741228 | 2.40 |
ENSDART00000112035
ENSDART00000127664 |
myf5
|
myogenic factor 5 |
| chr4_-_16451375 | 2.35 |
ENSDART00000192700
ENSDART00000128835 |
wu:fc23c09
|
wu:fc23c09 |
| chr6_-_36552844 | 2.33 |
ENSDART00000023613
|
her6
|
hairy-related 6 |
| chr23_-_21453614 | 2.28 |
ENSDART00000079274
|
her4.1
|
hairy-related 4, tandem duplicate 1 |
| chr19_+_42432625 | 2.24 |
ENSDART00000076938
|
pogza
|
pogo transposable element derived with ZNF domain a |
| chr8_-_36554675 | 2.23 |
ENSDART00000132804
ENSDART00000078746 |
ccdc157
|
coiled-coil domain containing 157 |
| chr2_-_32237916 | 2.08 |
ENSDART00000141418
|
fam49ba
|
family with sequence similarity 49, member Ba |
| chr20_+_14789148 | 2.06 |
ENSDART00000164761
|
tmed5
|
transmembrane p24 trafficking protein 5 |
| chr10_+_38512270 | 1.97 |
ENSDART00000109752
|
serpinh1a
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1a |
| chr16_+_40575742 | 1.95 |
ENSDART00000161503
|
ccne2
|
cyclin E2 |
| chr21_-_37194365 | 1.88 |
ENSDART00000100286
|
fgfr4
|
fibroblast growth factor receptor 4 |
| chr10_-_44306636 | 1.87 |
ENSDART00000191068
|
CDK2AP1
|
cyclin dependent kinase 2 associated protein 1 |
| chr24_+_18714212 | 1.84 |
ENSDART00000171181
|
cspp1a
|
centrosome and spindle pole associated protein 1a |
| chr20_-_39596338 | 1.83 |
ENSDART00000023531
|
hey2
|
hes-related family bHLH transcription factor with YRPW motif 2 |
| chr1_-_51720633 | 1.82 |
ENSDART00000045894
|
rnaseh2a
|
ribonuclease H2, subunit A |
| chr14_-_4321874 | 1.76 |
ENSDART00000042672
|
guf1
|
GUF1 homolog, GTPase |
| chr16_+_9762261 | 1.73 |
ENSDART00000020654
|
psmd4b
|
proteasome 26S subunit, non-ATPase 4b |
| chr6_+_3716666 | 1.71 |
ENSDART00000041627
|
ssb
|
Sjogren syndrome antigen B (autoantigen La) |
| chr19_-_18127629 | 1.70 |
ENSDART00000187722
|
snx10a
|
sorting nexin 10a |
| chr17_+_16046132 | 1.59 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr19_-_18127808 | 1.59 |
ENSDART00000108627
|
snx10a
|
sorting nexin 10a |
| chr5_+_65040228 | 1.57 |
ENSDART00000164278
|
pmpca
|
peptidase (mitochondrial processing) alpha |
| chr6_+_45932276 | 1.56 |
ENSDART00000103491
|
rbp7b
|
retinol binding protein 7b, cellular |
| chr17_-_2573021 | 1.56 |
ENSDART00000074181
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr25_+_16646113 | 1.54 |
ENSDART00000110426
|
cecr2
|
cat eye syndrome chromosome region, candidate 2 |
| chr3_+_3641429 | 1.53 |
ENSDART00000092393
|
plbd1
|
phospholipase B domain containing 1 |
| chr5_-_36597612 | 1.52 |
ENSDART00000031270
ENSDART00000122098 |
rhogc
|
ras homolog gene family, member Gc |
| chr24_+_24170914 | 1.51 |
ENSDART00000127842
|
si:dkey-226l10.6
|
si:dkey-226l10.6 |
| chr25_+_20272145 | 1.48 |
ENSDART00000109605
|
si:dkey-219c3.2
|
si:dkey-219c3.2 |
| chr16_+_5579744 | 1.44 |
ENSDART00000147973
|
macf1b
|
microtubule-actin crosslinking factor 1 b |
| chr8_+_15277874 | 1.44 |
ENSDART00000146965
|
dnttip2
|
deoxynucleotidyltransferase, terminal, interacting protein 2 |
| chr23_+_17839187 | 1.43 |
ENSDART00000104647
|
prim1
|
DNA primase subunit 1 |
| chr21_+_45268112 | 1.39 |
ENSDART00000157136
|
tcf7
|
transcription factor 7 |
| chr15_+_9861973 | 1.36 |
ENSDART00000170945
|
si:dkey-13m3.2
|
si:dkey-13m3.2 |
| chr2_-_48539673 | 1.33 |
ENSDART00000168202
|
CR391991.4
|
|
| chr7_-_41726657 | 1.32 |
ENSDART00000099121
|
arl8
|
ADP-ribosylation factor-like 8 |
| chr21_+_26536950 | 1.30 |
ENSDART00000146315
|
stx5al
|
syntaxin 5A, like |
| chr15_-_6863317 | 1.30 |
ENSDART00000155128
ENSDART00000018945 |
meis3
|
myeloid ecotropic viral integration site 3 |
| chr25_+_2263857 | 1.29 |
ENSDART00000076439
|
yars2
|
tyrosyl-tRNA synthetase 2, mitochondrial |
| chr16_-_31718013 | 1.29 |
ENSDART00000190716
|
rbp5
|
retinol binding protein 1a, cellular |
| chr23_-_9855627 | 1.29 |
ENSDART00000180159
|
prkcbp1l
|
protein kinase C binding protein 1, like |
| chr10_+_36441124 | 1.25 |
ENSDART00000185626
|
uspl1
|
ubiquitin specific peptidase like 1 |
| chr20_+_14789305 | 1.25 |
ENSDART00000002463
|
tmed5
|
transmembrane p24 trafficking protein 5 |
| chr16_-_31717851 | 1.24 |
ENSDART00000169109
|
rbp5
|
retinol binding protein 1a, cellular |
| chr21_+_43172506 | 1.23 |
ENSDART00000121725
|
zcchc10
|
zinc finger, CCHC domain containing 10 |
| chr3_+_31058464 | 1.23 |
ENSDART00000153381
|
si:dkey-66i24.7
|
si:dkey-66i24.7 |
| chr4_+_45441669 | 1.22 |
ENSDART00000150745
|
si:ch211-162i8.7
|
si:ch211-162i8.7 |
| chr10_-_44306399 | 1.20 |
ENSDART00000180042
|
CDK2AP1
|
cyclin dependent kinase 2 associated protein 1 |
| chr22_-_26274177 | 1.20 |
ENSDART00000060978
|
wdr83
|
WD repeat domain containing 83 |
| chr4_+_9030609 | 1.19 |
ENSDART00000154399
|
aldh1l2
|
aldehyde dehydrogenase 1 family, member L2 |
| chr21_+_39673157 | 1.18 |
ENSDART00000100240
|
mrm3b
|
mitochondrial rRNA methyltransferase 3b |
| chr25_+_34915576 | 1.17 |
ENSDART00000073441
|
sntb2
|
syntrophin, beta 2 |
| chr3_-_3428938 | 1.16 |
ENSDART00000179811
ENSDART00000115282 ENSDART00000192263 ENSDART00000046454 |
A2ML1 (1 of many)
|
zgc:171445 |
| chr18_-_18587745 | 1.15 |
ENSDART00000191973
|
sf3b3
|
splicing factor 3b, subunit 3 |
| chr24_-_21090447 | 1.14 |
ENSDART00000136507
ENSDART00000140786 ENSDART00000184841 |
qtrt2
|
queuine tRNA-ribosyltransferase accessory subunit 2 |
| chr18_-_21746421 | 1.12 |
ENSDART00000188809
|
pskh1
|
protein serine kinase H1 |
| chr19_+_48018802 | 1.11 |
ENSDART00000161339
ENSDART00000166978 |
UBE2M
|
si:ch1073-205c8.3 |
| chr11_-_3535537 | 1.09 |
ENSDART00000165329
ENSDART00000009788 |
ddx19
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 19 (DBP5 homolog, yeast) |
| chr4_-_41269844 | 1.08 |
ENSDART00000186177
|
CR388165.2
|
|
| chr24_-_20658446 | 1.08 |
ENSDART00000127923
|
nktr
|
natural killer cell triggering receptor |
| chr13_+_21919786 | 1.07 |
ENSDART00000182440
|
ndst2a
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 2a |
| chr23_+_1730663 | 1.07 |
ENSDART00000149545
|
tgm1
|
transglutaminase 1, K polypeptide |
| chr17_+_5061135 | 1.05 |
ENSDART00000064313
|
cdc5l
|
CDC5 cell division cycle 5-like (S. pombe) |
| chr23_-_5783421 | 1.05 |
ENSDART00000131521
ENSDART00000019455 |
csrp1a
|
cysteine and glycine-rich protein 1a |
| chr7_+_15329819 | 1.05 |
ENSDART00000006018
|
mespaa
|
mesoderm posterior aa |
| chr16_-_32233463 | 1.04 |
ENSDART00000102016
|
calhm6
|
calcium homeostasis modulator family member 6 |
| chr22_-_506522 | 1.03 |
ENSDART00000106645
ENSDART00000067637 |
dstyk
|
dual serine/threonine and tyrosine protein kinase |
| chr1_+_17593392 | 1.02 |
ENSDART00000078889
|
helt
|
helt bHLH transcription factor |
| chr4_+_40329524 | 1.02 |
ENSDART00000165722
|
znf993
|
zinc finger protein 993 |
| chr1_+_1805294 | 1.01 |
ENSDART00000103850
|
atp1a1a.3
|
ATPase Na+/K+ transporting subunit alpha 1a, tandem duplicate 3 |
| chr22_-_3255341 | 1.00 |
ENSDART00000114232
|
gpr35.1
|
G protein-coupled receptor 35, tandem duplicate 1 |
| chr6_-_52235118 | 0.99 |
ENSDART00000191243
|
tomm34
|
translocase of outer mitochondrial membrane 34 |
| chr21_-_27213166 | 0.99 |
ENSDART00000146959
|
mark2a
|
MAP/microtubule affinity-regulating kinase 2a |
| chr16_-_24605969 | 0.98 |
ENSDART00000163305
ENSDART00000167121 |
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr6_-_30683637 | 0.98 |
ENSDART00000065212
|
ttc4
|
tetratricopeptide repeat domain 4 |
| chr7_+_54475134 | 0.96 |
ENSDART00000164063
|
zgc:153993
|
zgc:153993 |
| chr8_-_39654669 | 0.96 |
ENSDART00000145677
|
si:dkey-63d15.12
|
si:dkey-63d15.12 |
| chr14_+_743346 | 0.95 |
ENSDART00000110511
|
klb
|
klotho beta |
| chr7_-_41338923 | 0.95 |
ENSDART00000099138
|
ncf2
|
neutrophil cytosolic factor 2 |
| chr4_-_61406364 | 0.95 |
ENSDART00000142592
|
znf1021
|
zinc finger protein 1021 |
| chr17_-_45387134 | 0.94 |
ENSDART00000010975
|
tmem206
|
transmembrane protein 206 |
| chr5_+_9246458 | 0.94 |
ENSDART00000081772
|
susd1
|
sushi domain containing 1 |
| chr1_-_54100988 | 0.92 |
ENSDART00000192662
|
rfx1b
|
regulatory factor X, 1b (influences HLA class II expression) |
| chr2_-_20923864 | 0.91 |
ENSDART00000006870
|
ptgs2a
|
prostaglandin-endoperoxide synthase 2a |
| chr25_+_245018 | 0.91 |
ENSDART00000155344
|
zgc:92481
|
zgc:92481 |
| chr17_+_43659940 | 0.90 |
ENSDART00000145738
ENSDART00000075619 |
adob
|
2-aminoethanethiol (cysteamine) dioxygenase b |
| chr18_-_46369516 | 0.90 |
ENSDART00000018163
|
irf2bp1
|
interferon regulatory factor 2 binding protein 1 |
| chr20_-_3319642 | 0.89 |
ENSDART00000186743
ENSDART00000123096 |
marcksa
|
myristoylated alanine-rich protein kinase C substrate a |
| chr25_+_245438 | 0.89 |
ENSDART00000004689
|
zgc:92481
|
zgc:92481 |
| chr15_-_23508214 | 0.88 |
ENSDART00000115051
|
abcg4b
|
ATP-binding cassette, sub-family G (WHITE), member 4b |
| chr18_+_18612388 | 0.88 |
ENSDART00000186455
|
st3gal2
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 2 |
| chr16_-_27161410 | 0.88 |
ENSDART00000177503
|
LO017771.1
|
|
| chr6_+_21005725 | 0.87 |
ENSDART00000041370
|
cx44.2
|
connexin 44.2 |
| chr5_+_20319519 | 0.86 |
ENSDART00000004217
|
coro1ca
|
coronin, actin binding protein, 1Ca |
| chr21_-_32082130 | 0.85 |
ENSDART00000003978
ENSDART00000182050 |
mat2b
|
methionine adenosyltransferase II, beta |
| chr25_+_34915762 | 0.85 |
ENSDART00000191776
|
sntb2
|
syntrophin, beta 2 |
| chr8_+_36554816 | 0.85 |
ENSDART00000126687
|
sf3a1
|
splicing factor 3a, subunit 1 |
| chr23_-_14216506 | 0.85 |
ENSDART00000019620
|
ddx23
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 23 |
| chr1_+_57348756 | 0.85 |
ENSDART00000063750
|
b3gntl1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase-like 1 |
| chr7_+_49681040 | 0.83 |
ENSDART00000176372
ENSDART00000192172 |
rassf7b
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7b |
| chr15_-_6863150 | 0.82 |
ENSDART00000153815
|
meis3
|
myeloid ecotropic viral integration site 3 |
| chr18_-_17075098 | 0.82 |
ENSDART00000042496
ENSDART00000192284 ENSDART00000180307 |
tango6
|
transport and golgi organization 6 homolog (Drosophila) |
| chr20_-_9199721 | 0.82 |
ENSDART00000064140
|
ylpm1
|
YLP motif containing 1 |
| chr17_+_32374876 | 0.81 |
ENSDART00000183851
|
ywhaqb
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide b |
| chr5_+_4533244 | 0.81 |
ENSDART00000158826
|
CABZ01058650.1
|
Danio rerio thiosulfate sulfurtransferase/rhodanese-like domain-containing protein 1 (LOC561325), mRNA. |
| chr8_+_23147609 | 0.78 |
ENSDART00000180284
|
gid8a
|
GID complex subunit 8 homolog a (S. cerevisiae) |
| chr9_+_50175366 | 0.78 |
ENSDART00000170352
|
cobll1b
|
cordon-bleu WH2 repeat protein-like 1b |
| chr11_-_20956309 | 0.78 |
ENSDART00000188659
|
CABZ01008739.1
|
|
| chr16_+_26449615 | 0.78 |
ENSDART00000039746
|
epb41b
|
erythrocyte membrane protein band 4.1b |
| chr14_+_109016 | 0.77 |
ENSDART00000158405
|
gpc2
|
glypican 2 |
| chr21_+_20549395 | 0.77 |
ENSDART00000181633
|
efna5a
|
ephrin-A5a |
| chr17_+_11372531 | 0.76 |
ENSDART00000130975
ENSDART00000149366 |
timm9
|
translocase of inner mitochondrial membrane 9 homolog |
| chr20_+_53368611 | 0.75 |
ENSDART00000060432
|
cdc40
|
cell division cycle 40 homolog (S. cerevisiae) |
| chr13_-_34781984 | 0.75 |
ENSDART00000172138
|
ism1
|
isthmin 1 |
| chr3_-_7436932 | 0.72 |
ENSDART00000163501
|
CR847850.1
|
|
| chr2_+_8112449 | 0.72 |
ENSDART00000138136
|
chst2a
|
carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2a |
| chr19_+_3842891 | 0.70 |
ENSDART00000159043
|
lsm10
|
LSM10, U7 small nuclear RNA associated |
| chr6_-_55423220 | 0.70 |
ENSDART00000158929
|
ctsa
|
cathepsin A |
| chr2_-_38287987 | 0.68 |
ENSDART00000185329
ENSDART00000061677 |
si:ch211-14a17.6
|
si:ch211-14a17.6 |
| chr16_+_2905150 | 0.68 |
ENSDART00000109980
|
lars2
|
leucyl-tRNA synthetase 2, mitochondrial |
| chr4_-_60518031 | 0.68 |
ENSDART00000157489
|
si:dkey-211i20.4
|
si:dkey-211i20.4 |
| chr22_+_567733 | 0.67 |
ENSDART00000171036
|
usp49
|
ubiquitin specific peptidase 49 |
| chr14_+_13454840 | 0.67 |
ENSDART00000161854
|
pls3
|
plastin 3 (T isoform) |
| chr12_-_4331117 | 0.67 |
ENSDART00000008893
|
ca15a
|
carbonic anhydrase XVa |
| chr7_+_27834130 | 0.66 |
ENSDART00000052656
|
rras2
|
RAS related 2 |
| chr24_+_39027481 | 0.65 |
ENSDART00000085565
|
capn15
|
calpain 15 |
| chr19_+_48018464 | 0.65 |
ENSDART00000172307
ENSDART00000163848 |
UBE2M
|
si:ch1073-205c8.3 |
| chr17_+_24851951 | 0.64 |
ENSDART00000180746
|
cx35.4
|
connexin 35.4 |
| chr5_-_29750377 | 0.63 |
ENSDART00000051474
|
barhl1a
|
BarH-like homeobox 1a |
| chr19_-_31802296 | 0.63 |
ENSDART00000103640
|
hey1
|
hes-related family bHLH transcription factor with YRPW motif 1 |
| chr5_+_41143563 | 0.63 |
ENSDART00000011229
|
sub1b
|
SUB1 homolog, transcriptional regulator b |
| chr22_+_25049563 | 0.63 |
ENSDART00000078173
|
dzank1
|
double zinc ribbon and ankyrin repeat domains 1 |
| chr17_+_6956696 | 0.63 |
ENSDART00000171368
|
zgc:172341
|
zgc:172341 |
| chr19_-_42462491 | 0.62 |
ENSDART00000131715
|
psmb4
|
proteasome subunit beta 4 |
| chr24_-_9989634 | 0.62 |
ENSDART00000115275
|
zgc:152652
|
zgc:152652 |
| chr5_-_22082918 | 0.60 |
ENSDART00000020908
|
zc4h2
|
zinc finger, C4H2 domain containing |
| chr6_+_58522557 | 0.60 |
ENSDART00000128062
|
arfrp1
|
ADP-ribosylation factor related protein 1 |
| chr22_+_30335936 | 0.59 |
ENSDART00000059923
|
mxi1
|
max interactor 1, dimerization protein |
| chr12_+_25223843 | 0.58 |
ENSDART00000077180
ENSDART00000127454 ENSDART00000122665 |
mta3
|
metastasis associated 1 family, member 3 |
| chr2_-_38282079 | 0.58 |
ENSDART00000145808
|
rnf212b
|
si:ch211-10e2.1 |
| chr3_-_41995321 | 0.57 |
ENSDART00000192277
|
ttyh3a
|
tweety family member 3a |
| chr7_-_17712665 | 0.57 |
ENSDART00000149047
|
men1
|
multiple endocrine neoplasia I |
| chr24_+_20658942 | 0.56 |
ENSDART00000142848
|
sec22c
|
SEC22 homolog C, vesicle trafficking protein |
| chr18_-_21640389 | 0.56 |
ENSDART00000100857
|
slc38a8a
|
solute carrier family 38, member 8a |
| chr20_+_218886 | 0.55 |
ENSDART00000002661
|
lama4
|
laminin, alpha 4 |
| chr24_+_20658760 | 0.53 |
ENSDART00000188362
|
sec22c
|
SEC22 homolog C, vesicle trafficking protein |
| chr12_-_37449396 | 0.53 |
ENSDART00000152951
|
cdc42ep4b
|
CDC42 effector protein (Rho GTPase binding) 4b |
| chr22_+_9049179 | 0.51 |
ENSDART00000192625
|
si:ch211-213a13.2
|
si:ch211-213a13.2 |
| chr4_-_5595237 | 0.51 |
ENSDART00000109854
|
vegfab
|
vascular endothelial growth factor Ab |
| chr7_+_24496894 | 0.51 |
ENSDART00000149994
|
nelfa
|
negative elongation factor complex member A |
| chr8_+_23147218 | 0.50 |
ENSDART00000030920
ENSDART00000141175 ENSDART00000146264 |
gid8a
|
GID complex subunit 8 homolog a (S. cerevisiae) |
| chr10_-_38468847 | 0.50 |
ENSDART00000133914
|
gdpd5a
|
glycerophosphodiester phosphodiesterase domain containing 5a |
| chr11_-_23322182 | 0.49 |
ENSDART00000111289
|
kiss1
|
KiSS-1 metastasis-suppressor |
| chr13_+_674351 | 0.48 |
ENSDART00000111328
|
cutc
|
cutC copper transporter homolog (E. coli) |
| chr3_-_36272670 | 0.48 |
ENSDART00000141638
|
prkar1aa
|
protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1) a |
| chr22_+_10752787 | 0.47 |
ENSDART00000186542
|
lsm7
|
LSM7 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr8_+_12925385 | 0.47 |
ENSDART00000085377
|
KIF2A
|
zgc:103670 |
| chr3_-_31158382 | 0.46 |
ENSDART00000076764
ENSDART00000076796 |
smg1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr3_-_3439150 | 0.46 |
ENSDART00000021286
|
A2ML1 (1 of many)
|
si:dkey-46g23.5 |
| chr5_-_37997774 | 0.44 |
ENSDART00000139616
ENSDART00000167694 |
si:dkey-111e8.1
|
si:dkey-111e8.1 |
| chr14_-_33521071 | 0.43 |
ENSDART00000052789
|
c1galt1c1
|
C1GALT1-specific chaperone 1 |
| chr9_+_23124252 | 0.42 |
ENSDART00000192377
|
BX936418.1
|
|
| chr20_-_54075136 | 0.42 |
ENSDART00000074255
|
mgat2
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr11_-_16394971 | 0.40 |
ENSDART00000180981
ENSDART00000179925 |
lrig1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr8_+_16726386 | 0.40 |
ENSDART00000144621
|
smim15
|
small integral membrane protein 15 |
| chr7_+_15324830 | 0.40 |
ENSDART00000189088
|
mespaa
|
mesoderm posterior aa |
| chr10_+_31951338 | 0.39 |
ENSDART00000019416
|
lhfpl6
|
LHFPL tetraspan subfamily member 6 |
| chr22_+_21965148 | 0.38 |
ENSDART00000136020
|
si:ch211-207c7.2
|
si:ch211-207c7.2 |
| chr4_-_74367912 | 0.38 |
ENSDART00000174199
ENSDART00000165257 |
ptprb
|
protein tyrosine phosphatase, receptor type, b |
| chr6_+_58522738 | 0.38 |
ENSDART00000157327
|
arfrp1
|
ADP-ribosylation factor related protein 1 |
| chr17_-_23416897 | 0.37 |
ENSDART00000163391
|
si:ch211-149k12.3
|
si:ch211-149k12.3 |
| chr14_+_26229056 | 0.36 |
ENSDART00000179045
|
LO018208.1
|
|
| chr8_-_46386024 | 0.34 |
ENSDART00000136602
ENSDART00000060919 ENSDART00000137472 |
qars
|
glutaminyl-tRNA synthetase |
| chr16_-_17300030 | 0.34 |
ENSDART00000149267
|
kel
|
Kell blood group, metallo-endopeptidase |
| chr3_+_17616201 | 0.34 |
ENSDART00000156775
|
rab5c
|
RAB5C, member RAS oncogene family |
| chr8_+_26292560 | 0.33 |
ENSDART00000053463
|
mgll
|
monoglyceride lipase |
| chr11_+_15480695 | 0.31 |
ENSDART00000104111
|
cdk5rap1
|
CDK5 regulatory subunit associated protein 1 |
| chr23_-_478201 | 0.31 |
ENSDART00000140749
|
si:ch73-181d5.4
|
si:ch73-181d5.4 |
| chr14_+_29775602 | 0.31 |
ENSDART00000125589
|
zgc:153146
|
zgc:153146 |
| chr7_-_43787616 | 0.30 |
ENSDART00000179758
|
cdh11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr3_-_53092509 | 0.28 |
ENSDART00000062081
|
lpar2a
|
lysophosphatidic acid receptor 2a |
| chr18_-_5209258 | 0.28 |
ENSDART00000183109
|
CABZ01080601.1
|
|
| chr2_-_59247811 | 0.27 |
ENSDART00000141384
|
ftr32
|
finTRIM family, member 32 |
| chr22_-_164944 | 0.27 |
ENSDART00000145379
|
fblim1
|
filamin binding LIM protein 1 |
| chr3_-_36750068 | 0.26 |
ENSDART00000173388
|
abcc6b.1
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 6b, tandem duplicate 1 |
| chr18_-_18937485 | 0.26 |
ENSDART00000139015
|
si:dkey-73n10.1
|
si:dkey-73n10.1 |
| chr13_-_50546634 | 0.25 |
ENSDART00000192127
|
CU570781.2
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of rbpja
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.7 | 4.1 | GO:0055016 | hypochord development(GO:0055016) |
| 0.7 | 3.3 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.7 | 4.6 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 0.6 | 1.9 | GO:1904251 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.6 | 1.8 | GO:0060844 | arterial endothelial cell fate commitment(GO:0060844) |
| 0.5 | 2.4 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.4 | 2.1 | GO:1901166 | neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.4 | 1.2 | GO:0042560 | 10-formyltetrahydrofolate metabolic process(GO:0009256) 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.4 | 1.4 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.3 | 1.4 | GO:0033153 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.3 | 1.3 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.3 | 1.8 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.3 | 3.2 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.3 | 3.7 | GO:0045176 | asymmetric protein localization(GO:0008105) apical protein localization(GO:0045176) |
| 0.3 | 2.3 | GO:0021794 | thalamus development(GO:0021794) |
| 0.3 | 1.3 | GO:1904867 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) protein localization to nucleoplasm(GO:1990173) |
| 0.2 | 1.4 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.2 | 0.9 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.2 | 0.9 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.2 | 1.6 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 2.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 2.2 | GO:2000344 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 1.4 | GO:0048041 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) regulation of focal adhesion assembly(GO:0051893) regulation of cell-substrate junction assembly(GO:0090109) |
| 0.1 | 1.0 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.1 | 0.9 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 2.3 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 1.7 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 4.4 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 0.4 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.1 | 2.5 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.1 | 1.8 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 1.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 1.0 | GO:0001836 | release of cytochrome c from mitochondria(GO:0001836) |
| 0.1 | 2.2 | GO:0008345 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.1 | 2.6 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.1 | 2.0 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.1 | 1.1 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 1.0 | GO:0006626 | protein targeting to mitochondrion(GO:0006626) |
| 0.1 | 2.1 | GO:0050870 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.1 | 0.6 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.1 | 0.8 | GO:0032204 | regulation of telomere maintenance(GO:0032204) |
| 0.1 | 0.7 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.4 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 1.0 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.1 | 0.2 | GO:0019062 | virion attachment to host cell(GO:0019062) adhesion of symbiont to host(GO:0044406) adhesion of symbiont to host cell(GO:0044650) |
| 0.1 | 0.3 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 0.5 | GO:0035188 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.1 | 1.0 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 0.5 | GO:0031268 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.9 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.1 | 1.5 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.1 | 0.5 | GO:0097531 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 5.7 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 1.1 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.5 | GO:0055070 | copper ion homeostasis(GO:0055070) |
| 0.0 | 0.3 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.7 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.7 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 1.8 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 1.3 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 1.5 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 2.4 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.0 | 0.5 | GO:0034244 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.9 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.0 | 0.5 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 1.0 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 1.1 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 1.4 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.6 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.0 | 0.9 | GO:0090148 | membrane fission(GO:0090148) |
| 0.0 | 0.7 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.5 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.8 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 1.0 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 1.5 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.2 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 3.5 | GO:0007219 | Notch signaling pathway(GO:0007219) |
| 0.0 | 0.5 | GO:0033555 | multicellular organismal response to stress(GO:0033555) |
| 0.0 | 0.6 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.6 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.6 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.4 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 1.0 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 0.1 | GO:0046931 | pore complex assembly(GO:0046931) |
| 0.0 | 0.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 1.1 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 3.8 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.0 | 4.3 | GO:0000398 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 1.4 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.6 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.5 | 2.4 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.5 | 1.4 | GO:1990077 | primosome complex(GO:1990077) |
| 0.5 | 1.8 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.4 | 2.6 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.3 | 0.8 | GO:0031362 | intrinsic component of external side of plasma membrane(GO:0031233) anchored component of external side of plasma membrane(GO:0031362) |
| 0.2 | 2.4 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.2 | 0.9 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 1.5 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.1 | 1.1 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.1 | 3.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 1.7 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.1 | 0.9 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 1.1 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.1 | 2.0 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 1.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 3.7 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 2.9 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.1 | 1.5 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 1.4 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 0.5 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 4.1 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.1 | 0.5 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 0.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 1.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.6 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 1.8 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.6 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.1 | GO:0044216 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.6 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.6 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 2.1 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 1.8 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.9 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 2.0 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 5.8 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 3.3 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 1.0 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.8 | GO:0005759 | mitochondrial matrix(GO:0005759) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.7 | 4.6 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.4 | 1.2 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.3 | 1.8 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.3 | 2.5 | GO:0016918 | retinal binding(GO:0016918) |
| 0.3 | 1.3 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.2 | 2.4 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.2 | 1.1 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.2 | 0.9 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.2 | 2.4 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.1 | 1.9 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 1.6 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.1 | 0.7 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 1.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 2.6 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 0.5 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 4.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.7 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.4 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.1 | 0.9 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 1.0 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.9 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.7 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.1 | 1.0 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 2.0 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 1.5 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.1 | 0.5 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 3.0 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.1 | 1.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 1.0 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 0.6 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 1.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 1.4 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.6 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.0 | 1.0 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.4 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.0 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.7 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.6 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 17.4 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 0.6 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 1.5 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 1.4 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 1.9 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.7 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0016519 | gastric inhibitory peptide receptor activity(GO:0016519) |
| 0.0 | 0.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 1.0 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 1.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.3 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.7 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 1.2 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.0 | 3.3 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 1.8 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 3.7 | GO:0051015 | actin filament binding(GO:0051015) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 1.8 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 2.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 3.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 2.8 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.6 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.9 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.1 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.4 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.5 | PID PLK1 PATHWAY | PLK1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.2 | 1.9 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.2 | 3.8 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 1.4 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.1 | 2.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 0.9 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 4.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 1.7 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 1.0 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.1 | 2.0 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 2.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 0.7 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 2.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 1.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 0.9 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 0.8 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 2.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.3 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.9 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.6 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.8 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.7 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.9 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.6 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 1.6 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |