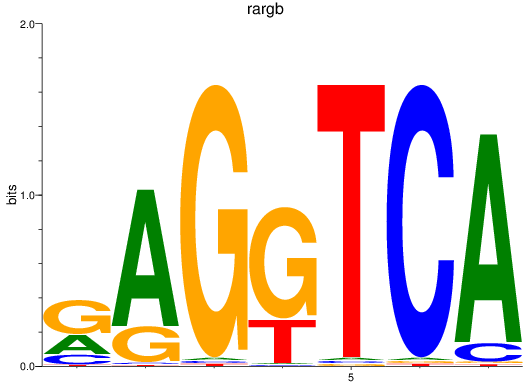
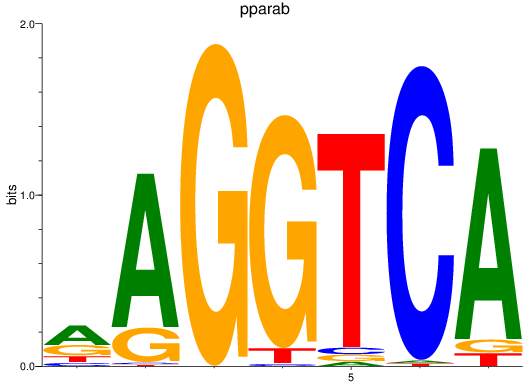
Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
Results for rargb_pparab
Z-value: 0.73
Transcription factors associated with rargb_pparab
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rargb
|
ENSDARG00000054003 | retinoic acid receptor, gamma b |
|
pparab
|
ENSDARG00000054323 | peroxisome proliferator-activated receptor alpha b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rargb | dr11_v1_chr11_+_2089461_2089461 | 0.53 | 2.0e-02 | Click! |
| pparab | dr11_v1_chr25_+_2229301_2229301 | 0.41 | 7.9e-02 | Click! |
Activity profile of rargb_pparab motif
Sorted Z-values of rargb_pparab motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_44027391 | 3.42 |
ENSDART00000145404
|
crybb1
|
crystallin, beta B1 |
| chr4_-_16853464 | 3.18 |
ENSDART00000125743
ENSDART00000164570 |
slc25a3a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3a |
| chr6_-_13783604 | 3.00 |
ENSDART00000149536
ENSDART00000041269 ENSDART00000150102 |
cryba2a
|
crystallin, beta A2a |
| chr16_+_12022543 | 2.79 |
ENSDART00000012673
|
gnb3a
|
guanine nucleotide binding protein (G protein), beta polypeptide 3a |
| chr23_-_45504991 | 2.43 |
ENSDART00000148761
|
col24a1
|
collagen type XXIV alpha 1 |
| chr12_-_4683325 | 2.37 |
ENSDART00000152771
|
si:ch211-255p10.3
|
si:ch211-255p10.3 |
| chr20_+_5564042 | 2.31 |
ENSDART00000090934
ENSDART00000127050 |
nrxn3b
|
neurexin 3b |
| chr11_+_30729745 | 2.28 |
ENSDART00000103270
|
slc22a7a
|
solute carrier family 22 (organic anion transporter), member 7a |
| chr17_-_12385308 | 2.23 |
ENSDART00000080927
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr16_+_26012569 | 2.19 |
ENSDART00000148846
|
prss59.1
|
protease, serine, 59, tandem duplicate 1 |
| chr3_-_32170850 | 2.17 |
ENSDART00000055307
ENSDART00000157366 |
tnnt1
|
troponin T type 1 (skeletal, slow) |
| chr19_+_10341738 | 2.17 |
ENSDART00000128721
|
rcvrn3
|
recoverin 3 |
| chr6_+_48618512 | 2.15 |
ENSDART00000111190
|
FAM19A3
|
si:dkey-238f9.1 |
| chr24_-_38374744 | 2.02 |
ENSDART00000007208
|
lrrc4bb
|
leucine rich repeat containing 4Bb |
| chr5_-_41831646 | 1.93 |
ENSDART00000134326
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr19_-_5135345 | 1.87 |
ENSDART00000151787
|
gnb3b
|
guanine nucleotide binding protein (G protein), beta polypeptide 3b |
| chr1_-_55810730 | 1.80 |
ENSDART00000100551
|
zgc:136908
|
zgc:136908 |
| chr16_-_560574 | 1.80 |
ENSDART00000148452
|
irx2a
|
iroquois homeobox 2a |
| chr7_-_24520866 | 1.65 |
ENSDART00000077039
|
faah2b
|
fatty acid amide hydrolase 2b |
| chr23_-_39636195 | 1.61 |
ENSDART00000144439
|
vwa1
|
von Willebrand factor A domain containing 1 |
| chr5_-_30620625 | 1.60 |
ENSDART00000098273
|
tcnl
|
transcobalamin like |
| chr6_-_40722200 | 1.60 |
ENSDART00000035101
|
kbtbd12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr10_-_11385155 | 1.59 |
ENSDART00000064214
|
plac8.1
|
placenta-specific 8, tandem duplicate 1 |
| chr21_-_41870029 | 1.58 |
ENSDART00000182035
|
endou2
|
endonuclease, polyU-specific 2 |
| chr7_+_35068036 | 1.53 |
ENSDART00000022139
|
zgc:136461
|
zgc:136461 |
| chr12_+_34896956 | 1.50 |
ENSDART00000055415
|
prph2a
|
peripherin 2a (retinal degeneration, slow) |
| chr2_+_38039857 | 1.48 |
ENSDART00000159951
|
casq1a
|
calsequestrin 1a |
| chr4_-_1360495 | 1.46 |
ENSDART00000164623
|
ptn
|
pleiotrophin |
| chr15_+_20403903 | 1.45 |
ENSDART00000134182
|
cox7a1
|
cytochrome c oxidase subunit VIIa polypeptide 1 (muscle) |
| chr5_-_46980651 | 1.41 |
ENSDART00000181022
ENSDART00000168038 |
edil3a
|
EGF-like repeats and discoidin I-like domains 3a |
| chr9_-_23944470 | 1.33 |
ENSDART00000138754
|
col6a3
|
collagen, type VI, alpha 3 |
| chr3_+_30257582 | 1.31 |
ENSDART00000159497
ENSDART00000103457 ENSDART00000121883 |
mybpc2a
|
myosin binding protein C, fast type a |
| chr8_+_48613040 | 1.30 |
ENSDART00000121432
|
nppa
|
natriuretic peptide A |
| chr11_+_36989696 | 1.29 |
ENSDART00000045888
|
tkta
|
transketolase a |
| chr15_+_47386939 | 1.29 |
ENSDART00000128224
|
FO904873.1
|
|
| chr2_+_42191592 | 1.28 |
ENSDART00000144716
|
cavin4a
|
caveolae associated protein 4a |
| chr3_-_41795917 | 1.26 |
ENSDART00000182662
|
grifin
|
galectin-related inter-fiber protein |
| chr2_-_24289641 | 1.26 |
ENSDART00000128784
ENSDART00000123565 ENSDART00000141922 ENSDART00000184550 ENSDART00000191469 |
myh7l
|
myosin heavy chain 7-like |
| chr19_-_6988837 | 1.24 |
ENSDART00000145741
ENSDART00000167640 |
znf384l
|
zinc finger protein 384 like |
| chr3_+_32142382 | 1.23 |
ENSDART00000133035
|
syt5a
|
synaptotagmin Va |
| chr16_+_11483811 | 1.18 |
ENSDART00000169012
ENSDART00000173042 |
grik5
|
glutamate receptor, ionotropic, kainate 5 |
| chr3_-_13147310 | 1.16 |
ENSDART00000160840
|
prkar1b
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr19_-_3193912 | 1.14 |
ENSDART00000133159
|
si:ch211-133n4.6
|
si:ch211-133n4.6 |
| chr10_-_29900546 | 1.13 |
ENSDART00000147441
|
lim2.1
|
lens intrinsic membrane protein 2.1 |
| chr6_-_31348999 | 1.13 |
ENSDART00000153734
|
dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr15_-_44512461 | 1.13 |
ENSDART00000155456
|
gria4a
|
glutamate receptor, ionotropic, AMPA 4a |
| chr20_+_30490682 | 1.12 |
ENSDART00000184871
|
myt1la
|
myelin transcription factor 1-like, a |
| chr8_+_24854600 | 1.12 |
ENSDART00000156570
|
slc6a17
|
solute carrier family 6 (neutral amino acid transporter), member 17 |
| chr21_+_39100289 | 1.08 |
ENSDART00000075958
|
slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr16_-_17175731 | 1.08 |
ENSDART00000183057
|
opn9
|
opsin 9 |
| chr17_-_31058900 | 1.06 |
ENSDART00000134998
ENSDART00000104307 ENSDART00000172721 |
eml1
|
echinoderm microtubule associated protein like 1 |
| chr6_-_49873020 | 1.06 |
ENSDART00000148511
|
gnas
|
GNAS complex locus |
| chr12_-_30841679 | 1.06 |
ENSDART00000105594
|
crygmx
|
crystallin, gamma MX |
| chr11_-_11471857 | 1.05 |
ENSDART00000030103
|
krt94
|
keratin 94 |
| chr2_+_28672152 | 1.05 |
ENSDART00000157410
ENSDART00000169614 |
nadsyn1
|
NAD synthetase 1 |
| chr5_+_1278092 | 1.04 |
ENSDART00000147972
ENSDART00000159783 |
dnm1a
|
dynamin 1a |
| chr5_-_68916455 | 1.03 |
ENSDART00000171465
|
ank1a
|
ankyrin 1, erythrocytic a |
| chr16_+_5774977 | 1.03 |
ENSDART00000134202
|
ccka
|
cholecystokinin a |
| chr11_-_17713987 | 1.02 |
ENSDART00000090401
|
fam19a4b
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A4b |
| chr19_-_703898 | 0.99 |
ENSDART00000181096
ENSDART00000121462 |
slc6a19a.2
|
solute carrier family 6 (neutral amino acid transporter), member 19a, tandem duplicate 2 |
| chr8_-_25329967 | 0.99 |
ENSDART00000139682
|
eps8l3b
|
EPS8-like 3b |
| chr25_+_29160102 | 0.99 |
ENSDART00000162854
|
pkmb
|
pyruvate kinase M1/2b |
| chr5_+_38276582 | 0.98 |
ENSDART00000158532
|
gnb2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr24_-_17047918 | 0.98 |
ENSDART00000020204
|
msrb2
|
methionine sulfoxide reductase B2 |
| chr6_+_3828560 | 0.97 |
ENSDART00000185273
ENSDART00000179091 |
gad1b
|
glutamate decarboxylase 1b |
| chr20_+_13969414 | 0.97 |
ENSDART00000049864
|
rd3
|
retinal degeneration 3 |
| chr11_+_11201096 | 0.96 |
ENSDART00000171916
ENSDART00000171521 ENSDART00000087105 ENSDART00000159603 |
myom2a
|
myomesin 2a |
| chr13_-_22843562 | 0.94 |
ENSDART00000142738
|
pbld
|
phenazine biosynthesis like protein domain containing |
| chr15_+_28096152 | 0.93 |
ENSDART00000100293
ENSDART00000140092 |
crybb1l3
|
crystallin, beta B1, like 3 |
| chr6_-_40722480 | 0.92 |
ENSDART00000188187
|
kbtbd12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr17_+_30448452 | 0.92 |
ENSDART00000153939
|
lpin1
|
lipin 1 |
| chr4_+_17280868 | 0.91 |
ENSDART00000145349
|
bcat1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr24_+_29449690 | 0.91 |
ENSDART00000105743
ENSDART00000193556 ENSDART00000145816 |
ntng1a
|
netrin g1a |
| chr19_+_43314700 | 0.90 |
ENSDART00000049045
ENSDART00000133158 |
matn1
|
matrilin 1 |
| chr3_+_13440900 | 0.90 |
ENSDART00000143715
|
GNG14
|
si:dkey-117i10.1 |
| chr23_-_11870962 | 0.90 |
ENSDART00000143481
|
si:dkey-178k16.1
|
si:dkey-178k16.1 |
| chr8_+_23142946 | 0.89 |
ENSDART00000152933
|
si:ch211-196c10.13
|
si:ch211-196c10.13 |
| chr18_-_16179129 | 0.86 |
ENSDART00000125353
|
slc6a15
|
solute carrier family 6 (neutral amino acid transporter), member 15 |
| chr15_+_45563656 | 0.85 |
ENSDART00000157501
|
cldn15lb
|
claudin 15-like b |
| chr6_+_10450000 | 0.84 |
ENSDART00000151288
ENSDART00000187431 ENSDART00000192474 ENSDART00000188214 ENSDART00000184766 ENSDART00000190082 |
kcnh7
|
potassium channel, voltage gated eag related subfamily H, member 7 |
| chr18_+_8340886 | 0.84 |
ENSDART00000081132
|
cpt1b
|
carnitine palmitoyltransferase 1B (muscle) |
| chr3_+_12710350 | 0.84 |
ENSDART00000157959
|
cyp2k18
|
cytochrome P450, family 2, subfamily K, polypeptide 18 |
| chr5_+_42467867 | 0.83 |
ENSDART00000172028
|
pimr58
|
Pim proto-oncogene, serine/threonine kinase, related 58 |
| chr11_+_45421761 | 0.82 |
ENSDART00000167347
|
hrasls
|
HRAS-like suppressor |
| chr5_-_55395964 | 0.82 |
ENSDART00000145791
|
prune2
|
prune homolog 2 (Drosophila) |
| chr17_-_40397752 | 0.82 |
ENSDART00000178483
|
BX548062.1
|
|
| chr7_+_568819 | 0.82 |
ENSDART00000173716
|
nrxn2b
|
neurexin 2b |
| chr5_-_10768258 | 0.82 |
ENSDART00000157043
|
rtn4r
|
reticulon 4 receptor |
| chr18_+_43365890 | 0.81 |
ENSDART00000173113
|
si:ch211-129p13.1
|
si:ch211-129p13.1 |
| chr24_+_39137001 | 0.80 |
ENSDART00000181086
ENSDART00000183724 ENSDART00000193466 |
tbc1d24
|
TBC1 domain family, member 24 |
| chr13_+_22480496 | 0.80 |
ENSDART00000136863
ENSDART00000131870 ENSDART00000078720 ENSDART00000078740 ENSDART00000139218 |
ldb3a
|
LIM domain binding 3a |
| chr23_-_27571667 | 0.80 |
ENSDART00000008174
|
pfkma
|
phosphofructokinase, muscle a |
| chr10_-_30785501 | 0.78 |
ENSDART00000020054
|
opcml
|
opioid binding protein/cell adhesion molecule-like |
| chr2_+_7818368 | 0.78 |
ENSDART00000007068
|
kcnmb2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr14_-_14659023 | 0.77 |
ENSDART00000170355
ENSDART00000159888 ENSDART00000172241 |
nsdhl
|
NAD(P) dependent steroid dehydrogenase-like |
| chr18_+_16330025 | 0.77 |
ENSDART00000142353
|
nts
|
neurotensin |
| chr5_+_36974931 | 0.77 |
ENSDART00000193063
|
gjd1a
|
gap junction protein delta 1a |
| chr19_-_6631900 | 0.76 |
ENSDART00000144571
|
pvrl2l
|
poliovirus receptor-related 2 like |
| chr15_-_28618502 | 0.76 |
ENSDART00000086902
|
slc6a4a
|
solute carrier family 6 (neurotransmitter transporter), member 4a |
| chr12_-_47782623 | 0.76 |
ENSDART00000115742
|
selenou1b
|
selenoprotein U1b |
| chr7_+_50109239 | 0.76 |
ENSDART00000021605
|
LRRC4C (1 of many)
|
si:dkey-6l15.1 |
| chr20_-_25709247 | 0.75 |
ENSDART00000146711
|
si:dkeyp-117h8.2
|
si:dkeyp-117h8.2 |
| chr5_-_41838354 | 0.75 |
ENSDART00000146793
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr22_+_19290199 | 0.74 |
ENSDART00000148173
|
si:dkey-21e2.15
|
si:dkey-21e2.15 |
| chr13_-_18691041 | 0.74 |
ENSDART00000057867
|
sfxn3
|
sideroflexin 3 |
| chr7_-_28413224 | 0.74 |
ENSDART00000076502
|
rerglb
|
RERG/RAS-like b |
| chr8_-_46700278 | 0.74 |
ENSDART00000143780
|
gpr153
|
G protein-coupled receptor 153 |
| chr16_-_17162485 | 0.73 |
ENSDART00000123011
|
iffo1b
|
intermediate filament family orphan 1b |
| chr15_+_45994123 | 0.73 |
ENSDART00000124704
|
lrfn1
|
leucine rich repeat and fibronectin type III domain containing 1 |
| chr5_-_23280098 | 0.73 |
ENSDART00000126540
ENSDART00000051533 |
plp1b
|
proteolipid protein 1b |
| chr14_+_21699414 | 0.72 |
ENSDART00000169942
|
stx3a
|
syntaxin 3A |
| chr21_-_22724980 | 0.72 |
ENSDART00000035469
|
c1qa
|
complement component 1, q subcomponent, A chain |
| chr11_+_25477643 | 0.71 |
ENSDART00000065941
|
opn1lw1
|
opsin 1 (cone pigments), long-wave-sensitive, 1 |
| chr23_-_35756115 | 0.71 |
ENSDART00000043429
|
jph2
|
junctophilin 2 |
| chr13_-_22862133 | 0.71 |
ENSDART00000138563
|
pbld2
|
phenazine biosynthesis-like protein domain containing 2 |
| chr23_-_32129569 | 0.70 |
ENSDART00000167761
ENSDART00000139569 |
zgc:92658
|
zgc:92658 |
| chr25_+_16945348 | 0.70 |
ENSDART00000016591
|
fgf6a
|
fibroblast growth factor 6a |
| chr9_-_49493305 | 0.69 |
ENSDART00000148707
ENSDART00000148561 |
xirp2b
|
xin actin binding repeat containing 2b |
| chr1_-_50859053 | 0.69 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr16_+_31804590 | 0.69 |
ENSDART00000167321
|
wnt4b
|
wingless-type MMTV integration site family, member 4b |
| chr23_+_44741500 | 0.69 |
ENSDART00000166421
|
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr11_+_36243774 | 0.69 |
ENSDART00000023323
|
zgc:172270
|
zgc:172270 |
| chr17_+_22381215 | 0.68 |
ENSDART00000162670
ENSDART00000128875 |
slc8a1b
|
solute carrier family 8 (sodium/calcium exchanger), member 1b |
| chr15_+_45575180 | 0.68 |
ENSDART00000180441
|
cldn15lb
|
claudin 15-like b |
| chr1_+_58094551 | 0.67 |
ENSDART00000146316
|
si:ch211-114l13.1
|
si:ch211-114l13.1 |
| chr2_-_5728843 | 0.67 |
ENSDART00000014020
|
sst2
|
somatostatin 2 |
| chr9_-_48736388 | 0.67 |
ENSDART00000022074
|
dhrs9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr16_+_29650698 | 0.67 |
ENSDART00000137153
|
tmod4
|
tropomodulin 4 (muscle) |
| chr10_+_26834985 | 0.67 |
ENSDART00000147518
|
fam89b
|
family with sequence similarity 89, member B |
| chr3_-_5829501 | 0.66 |
ENSDART00000091017
|
pkn1b
|
protein kinase N1b |
| chr17_+_25290136 | 0.65 |
ENSDART00000173295
|
kbtbd11
|
kelch repeat and BTB (POZ) domain containing 11 |
| chr7_-_48805181 | 0.65 |
ENSDART00000015884
|
mfge8a
|
milk fat globule-EGF factor 8 protein a |
| chr17_-_7818944 | 0.65 |
ENSDART00000135538
ENSDART00000037541 |
rmnd1
|
required for meiotic nuclear division 1 homolog |
| chr18_+_38321039 | 0.65 |
ENSDART00000132534
ENSDART00000111260 ENSDART00000192806 |
alx4b
|
ALX homeobox 4b |
| chr13_+_22480857 | 0.65 |
ENSDART00000078721
ENSDART00000044719 ENSDART00000130957 ENSDART00000078757 ENSDART00000130424 ENSDART00000078747 |
ldb3a
|
LIM domain binding 3a |
| chr3_+_1015867 | 0.64 |
ENSDART00000109912
|
si:ch1073-464p5.5
|
si:ch1073-464p5.5 |
| chr20_-_1141722 | 0.64 |
ENSDART00000152675
|
gabrr1
|
gamma-aminobutyric acid (GABA) A receptor, rho 1 |
| chr12_+_46708920 | 0.64 |
ENSDART00000153089
|
exoc7
|
exocyst complex component 7 |
| chr25_-_19420949 | 0.64 |
ENSDART00000181338
|
map1ab
|
microtubule-associated protein 1Ab |
| chr8_+_47698098 | 0.63 |
ENSDART00000186662
|
CU499311.1
|
|
| chr6_-_27139396 | 0.63 |
ENSDART00000055848
|
zgc:103559
|
zgc:103559 |
| chr10_-_39153959 | 0.63 |
ENSDART00000150193
ENSDART00000111362 |
slc37a4b
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4b |
| chr1_+_29096881 | 0.62 |
ENSDART00000075539
|
cryaa
|
crystallin, alpha A |
| chr14_+_33413980 | 0.62 |
ENSDART00000052780
ENSDART00000124437 ENSDART00000173327 |
ndufa1
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 1 |
| chr20_-_8443425 | 0.62 |
ENSDART00000083908
|
dab1a
|
Dab, reelin signal transducer, homolog 1a (Drosophila) |
| chr8_-_25980694 | 0.62 |
ENSDART00000135456
|
si:dkey-72l14.3
|
si:dkey-72l14.3 |
| chr21_-_39546737 | 0.62 |
ENSDART00000006971
|
sept4a
|
septin 4a |
| chr11_+_30162407 | 0.62 |
ENSDART00000190333
ENSDART00000127502 |
cdkl5
|
cyclin-dependent kinase-like 5 |
| chr19_+_40856534 | 0.62 |
ENSDART00000051950
|
gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr6_-_46875310 | 0.62 |
ENSDART00000154442
|
igfn1.3
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 3 |
| chr11_+_5588122 | 0.62 |
ENSDART00000113281
|
zgc:172302
|
zgc:172302 |
| chr6_+_8176486 | 0.61 |
ENSDART00000193308
|
nfil3-5
|
nuclear factor, interleukin 3 regulated, member 5 |
| chr19_+_40861853 | 0.61 |
ENSDART00000126470
|
zgc:85777
|
zgc:85777 |
| chr12_+_7491690 | 0.61 |
ENSDART00000152564
|
phyhiplb
|
phytanoyl-CoA 2-hydroxylase interacting protein-like b |
| chr16_-_33095161 | 0.61 |
ENSDART00000187648
|
dopey1
|
dopey family member 1 |
| chr13_+_27314795 | 0.60 |
ENSDART00000128726
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr2_-_127945 | 0.60 |
ENSDART00000056453
|
igfbp1b
|
insulin-like growth factor binding protein 1b |
| chr7_+_36539124 | 0.60 |
ENSDART00000173653
|
chd9
|
chromodomain helicase DNA binding protein 9 |
| chr12_-_47601845 | 0.60 |
ENSDART00000169548
ENSDART00000182889 |
rgs7b
|
regulator of G protein signaling 7b |
| chr15_+_45640906 | 0.60 |
ENSDART00000149361
ENSDART00000149079 |
sagb
|
S-antigen; retina and pineal gland (arrestin) b |
| chr17_-_5583345 | 0.60 |
ENSDART00000035944
|
clic5a
|
chloride intracellular channel 5a |
| chr23_+_6232895 | 0.60 |
ENSDART00000139795
|
syt2a
|
synaptotagmin IIa |
| chr14_-_33454595 | 0.60 |
ENSDART00000109615
ENSDART00000173267 ENSDART00000185737 ENSDART00000190989 |
tmem255a
|
transmembrane protein 255A |
| chr17_-_6738538 | 0.60 |
ENSDART00000157125
|
vsnl1b
|
visinin-like 1b |
| chr6_-_28980756 | 0.59 |
ENSDART00000014661
|
glmnb
|
glomulin, FKBP associated protein b |
| chr19_+_41169996 | 0.59 |
ENSDART00000048438
|
asb4
|
ankyrin repeat and SOCS box containing 4 |
| chr8_-_53108207 | 0.59 |
ENSDART00000111023
|
b3galt4
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 4 |
| chr8_+_22851472 | 0.59 |
ENSDART00000041564
|
cacna1fb
|
calcium channel, voltage-dependent, L type, alpha 1F subunit |
| chr16_+_11724230 | 0.59 |
ENSDART00000060266
|
ceacam1
|
carcinoembryonic antigen-related cell adhesion molecule 1 |
| chr23_+_44611864 | 0.59 |
ENSDART00000145905
ENSDART00000132361 |
eno3
|
enolase 3, (beta, muscle) |
| chr20_-_43771871 | 0.59 |
ENSDART00000153304
|
matn3a
|
matrilin 3a |
| chr4_+_47736069 | 0.59 |
ENSDART00000193329
|
si:ch211-196f19.1
|
si:ch211-196f19.1 |
| chr10_-_35108683 | 0.59 |
ENSDART00000049633
|
zgc:110006
|
zgc:110006 |
| chr23_-_26535875 | 0.59 |
ENSDART00000135988
|
si:dkey-205h13.2
|
si:dkey-205h13.2 |
| chr25_-_204019 | 0.58 |
ENSDART00000188440
ENSDART00000191735 |
FP236318.2
|
|
| chr13_-_37254777 | 0.58 |
ENSDART00000139734
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr23_+_3616224 | 0.58 |
ENSDART00000190917
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr20_+_1996202 | 0.58 |
ENSDART00000184143
|
CABZ01092781.1
|
|
| chr15_+_19797918 | 0.58 |
ENSDART00000113314
|
si:ch211-229d2.5
|
si:ch211-229d2.5 |
| chr16_+_50289916 | 0.58 |
ENSDART00000168861
ENSDART00000167332 |
hamp
|
hepcidin antimicrobial peptide |
| chr22_+_20427170 | 0.57 |
ENSDART00000136744
|
foxq2
|
forkhead box Q2 |
| chr23_+_3594171 | 0.57 |
ENSDART00000159609
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr16_-_47483142 | 0.57 |
ENSDART00000147072
|
cthrc1b
|
collagen triple helix repeat containing 1b |
| chr10_+_40973524 | 0.57 |
ENSDART00000176660
ENSDART00000114966 |
antxr1b
|
anthrax toxin receptor 1b |
| chr7_-_29339761 | 0.57 |
ENSDART00000086664
|
trpm1a
|
transient receptor potential cation channel, subfamily M, member 1a |
| chr19_+_40856807 | 0.57 |
ENSDART00000139083
|
gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr21_+_25231160 | 0.56 |
ENSDART00000063089
ENSDART00000139127 |
gng8
|
guanine nucleotide binding protein (G protein), gamma 8 |
| chr8_-_4097722 | 0.56 |
ENSDART00000135006
|
cux2b
|
cut-like homeobox 2b |
| chr6_-_39344259 | 0.56 |
ENSDART00000104074
|
zgc:158846
|
zgc:158846 |
| chr4_-_4932619 | 0.56 |
ENSDART00000103293
|
ndufa5
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5 |
| chr11_+_13223625 | 0.56 |
ENSDART00000161275
|
abcb11b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11b |
| chr7_+_24520518 | 0.56 |
ENSDART00000173604
|
btr09
|
bloodthirsty-related gene family, member 9 |
| chr20_-_34090740 | 0.55 |
ENSDART00000062539
ENSDART00000008140 |
pdcb
|
phosducin b |
| chr14_+_17376940 | 0.55 |
ENSDART00000054590
ENSDART00000010148 |
spon2b
|
spondin 2b, extracellular matrix protein |
| chr4_-_27301356 | 0.55 |
ENSDART00000100444
|
fam19a5a
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5a |
| chr10_-_31782616 | 0.55 |
ENSDART00000128839
|
fez1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr10_+_7563755 | 0.55 |
ENSDART00000165877
|
purg
|
purine-rich element binding protein G |
| chr8_+_21406769 | 0.54 |
ENSDART00000135766
|
si:dkey-163f12.6
|
si:dkey-163f12.6 |
| chr23_-_4915118 | 0.54 |
ENSDART00000060714
|
atp6ap1a
|
ATPase H+ transporting accessory protein 1a |
| chr9_-_44905867 | 0.54 |
ENSDART00000138316
ENSDART00000131252 ENSDART00000179383 ENSDART00000159337 |
zgc:66484
|
zgc:66484 |
Network of associatons between targets according to the STRING database.
First level regulatory network of rargb_pparab
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0051228 | mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.6 | 2.3 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.5 | 1.5 | GO:0014809 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.4 | 1.3 | GO:1905072 | cardiac jelly development(GO:1905072) |
| 0.4 | 1.1 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.3 | 2.0 | GO:0038065 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.3 | 1.0 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.3 | 2.2 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.3 | 3.4 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.3 | 0.3 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.3 | 0.8 | GO:0009750 | response to fructose(GO:0009750) |
| 0.3 | 1.3 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.2 | 1.0 | GO:0030091 | protein repair(GO:0030091) |
| 0.2 | 0.7 | GO:0044108 | calcitriol biosynthetic process from calciol(GO:0036378) vitamin D biosynthetic process(GO:0042368) cellular alcohol metabolic process(GO:0044107) cellular alcohol biosynthetic process(GO:0044108) |
| 0.2 | 0.9 | GO:0045023 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.2 | 0.9 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.2 | 1.1 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.2 | 1.1 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.2 | 0.7 | GO:0070131 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.2 | 1.7 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) |
| 0.2 | 0.8 | GO:0006837 | serotonin transport(GO:0006837) serotonin uptake(GO:0051610) |
| 0.2 | 2.6 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.2 | 0.5 | GO:2000434 | regulation of protein neddylation(GO:2000434) |
| 0.2 | 1.0 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.1 | 3.4 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 0.7 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.1 | 0.6 | GO:0015722 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.1 | 0.4 | GO:0072679 | thymocyte migration(GO:0072679) |
| 0.1 | 0.4 | GO:0032757 | interleukin-8 production(GO:0032637) regulation of interleukin-8 production(GO:0032677) positive regulation of interleukin-8 production(GO:0032757) |
| 0.1 | 0.5 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 0.8 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 0.5 | GO:0070459 | copulation(GO:0007620) regulation of epinephrine secretion(GO:0014060) positive regulation of epinephrine secretion(GO:0032812) positive regulation of catecholamine secretion(GO:0033605) penile erection(GO:0043084) epinephrine transport(GO:0048241) epinephrine secretion(GO:0048242) regulation of penile erection(GO:0060405) positive regulation of penile erection(GO:0060406) prolactin secretion(GO:0070459) |
| 0.1 | 0.1 | GO:0007618 | mating(GO:0007618) |
| 0.1 | 1.4 | GO:0061621 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 0.2 | GO:1903170 | negative regulation of calcium ion transmembrane transporter activity(GO:1901020) negative regulation of voltage-gated calcium channel activity(GO:1901386) negative regulation of calcium ion transmembrane transport(GO:1903170) |
| 0.1 | 0.5 | GO:0006598 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.1 | 0.6 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.1 | 0.4 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.1 | 0.5 | GO:0045938 | regulation of endocrine process(GO:0044060) positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) endocrine hormone secretion(GO:0060986) |
| 0.1 | 0.7 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.1 | 0.3 | GO:0042560 | 10-formyltetrahydrofolate metabolic process(GO:0009256) 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.1 | 0.3 | GO:0019556 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.1 | 0.8 | GO:0046864 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.1 | 0.5 | GO:0032615 | interleukin-12 production(GO:0032615) regulation of interleukin-12 production(GO:0032655) |
| 0.1 | 0.6 | GO:0051705 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) multi-organism behavior(GO:0051705) |
| 0.1 | 0.4 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.1 | 2.2 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 1.4 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.1 | 0.9 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 0.5 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 0.5 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.3 | GO:0046184 | pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) aldehyde biosynthetic process(GO:0046184) |
| 0.1 | 0.8 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.4 | GO:0019482 | beta-alanine metabolic process(GO:0019482) beta-alanine biosynthetic process(GO:0019483) |
| 0.1 | 0.4 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.1 | 0.5 | GO:0035889 | otolith tethering(GO:0035889) |
| 0.1 | 0.3 | GO:0042245 | RNA repair(GO:0042245) |
| 0.1 | 0.3 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.1 | 0.9 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.1 | 0.4 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.1 | 1.8 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.2 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.1 | 0.6 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.1 | 0.3 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.1 | 0.5 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.3 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 0.6 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.1 | 0.4 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 0.3 | GO:0006867 | asparagine transport(GO:0006867) |
| 0.1 | 2.0 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.1 | 0.8 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.1 | 0.6 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 1.2 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 0.6 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 0.7 | GO:0001765 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.1 | 0.2 | GO:0010312 | detoxification of zinc ion(GO:0010312) stress response to zinc ion(GO:1990359) |
| 0.1 | 9.0 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.1 | 0.4 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.1 | 0.3 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 0.9 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 1.2 | GO:0039014 | pronephric nephron tubule epithelial cell differentiation(GO:0035778) cell differentiation involved in pronephros development(GO:0039014) nephron tubule epithelial cell differentiation(GO:0072160) |
| 0.1 | 0.4 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.1 | 0.1 | GO:0051926 | negative regulation of calcium ion transport(GO:0051926) |
| 0.1 | 0.6 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.1 | GO:0042745 | circadian sleep/wake cycle process(GO:0022410) circadian sleep/wake cycle(GO:0042745) |
| 0.1 | 0.2 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.1 | 0.5 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.1 | 0.8 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 0.2 | GO:0099548 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.1 | 0.2 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.1 | 0.2 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.1 | 0.8 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.1 | 0.3 | GO:0006833 | water transport(GO:0006833) |
| 0.1 | 0.5 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 2.8 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.1 | 1.2 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.1 | 1.9 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 | 0.6 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 0.6 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.1 | 0.2 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 0.2 | GO:1902410 | mitotic cytokinetic process(GO:1902410) mitotic cleavage furrow formation(GO:1903673) |
| 0.1 | 0.2 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 0.6 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 0.3 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.0 | 0.1 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.0 | 0.4 | GO:0046887 | positive regulation of hormone secretion(GO:0046887) |
| 0.0 | 1.2 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.3 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.0 | 0.4 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.3 | GO:1901490 | regulation of lymphangiogenesis(GO:1901490) |
| 0.0 | 0.2 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.0 | 1.0 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.2 | GO:0003161 | cardiac conduction system development(GO:0003161) negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.6 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 0.4 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.5 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.3 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.3 | GO:0035523 | protein K29-linked deubiquitination(GO:0035523) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 1.6 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.9 | GO:0043507 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.4 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.0 | 0.4 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 0.3 | GO:0090178 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.2 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) |
| 0.0 | 0.5 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.2 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.2 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.8 | GO:0015669 | gas transport(GO:0015669) |
| 0.0 | 0.1 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.0 | 0.2 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.3 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.3 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.0 | 0.4 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.0 | 0.5 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.7 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 1.0 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 1.8 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.4 | GO:0001964 | startle response(GO:0001964) |
| 0.0 | 0.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.1 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.0 | 3.4 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.4 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.0 | 0.3 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.4 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.0 | 0.1 | GO:0008207 | C21-steroid hormone metabolic process(GO:0008207) |
| 0.0 | 0.1 | GO:0035992 | tendon formation(GO:0035992) |
| 0.0 | 0.2 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.2 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.3 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 1.0 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.0 | 0.1 | GO:0021828 | substrate-dependent cell migration(GO:0006929) pallium development(GO:0021543) cerebral cortex cell migration(GO:0021795) cerebral cortex tangential migration(GO:0021800) cerebral cortex tangential migration using cell-axon interactions(GO:0021824) substrate-dependent cerebral cortex tangential migration(GO:0021825) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) cerebral cortex development(GO:0021987) |
| 0.0 | 0.1 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.1 | GO:0090579 | establishment of mitotic sister chromatid cohesion(GO:0034087) establishment of protein localization to chromosome(GO:0070199) establishment of protein localization to chromatin(GO:0071169) transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.0 | 0.7 | GO:0090148 | membrane fission(GO:0090148) |
| 0.0 | 2.1 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.1 | GO:0030858 | positive regulation of epithelial cell differentiation(GO:0030858) regulation of endothelial cell differentiation(GO:0045601) |
| 0.0 | 0.2 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.0 | 0.2 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.4 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.5 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.5 | GO:0033555 | multicellular organismal response to stress(GO:0033555) |
| 0.0 | 0.3 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.2 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.1 | GO:0048308 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.0 | 0.1 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.2 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.0 | 0.3 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0072149 | visceral serous pericardium development(GO:0061032) glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.0 | 0.5 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.5 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.7 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.1 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.0 | 0.3 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.4 | GO:0050930 | regulation of positive chemotaxis(GO:0050926) positive regulation of positive chemotaxis(GO:0050927) induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.7 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.0 | 0.3 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.0 | 0.4 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.4 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.2 | GO:0061333 | renal tubule morphogenesis(GO:0061333) nephron tubule morphogenesis(GO:0072078) nephron epithelium morphogenesis(GO:0072088) |
| 0.0 | 0.2 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.7 | GO:0046463 | triglyceride biosynthetic process(GO:0019432) neutral lipid biosynthetic process(GO:0046460) acylglycerol biosynthetic process(GO:0046463) |
| 0.0 | 0.4 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.2 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.8 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 1.1 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.0 | 0.1 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) |
| 0.0 | 0.3 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.0 | 0.3 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 0.2 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 3.3 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.0 | 0.3 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.2 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.1 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.0 | 0.1 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.5 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.2 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.0 | 0.1 | GO:0070508 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.0 | 0.6 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.1 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.3 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.2 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.1 | GO:0070292 | N-acylphosphatidylethanolamine metabolic process(GO:0070292) |
| 0.0 | 0.2 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.4 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 2.0 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 1.0 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.0 | 0.1 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.0 | 0.1 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.5 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.2 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.0 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.0 | 0.1 | GO:0006691 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.0 | 0.4 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.1 | GO:0032475 | otolith formation(GO:0032475) |
| 0.0 | 0.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.8 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.3 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.1 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 0.0 | GO:0002637 | regulation of immunoglobulin production(GO:0002637) positive regulation of immunoglobulin production(GO:0002639) |
| 0.0 | 2.1 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 0.0 | GO:0098529 | neuromuscular junction development, skeletal muscle fiber(GO:0098529) |
| 0.0 | 1.1 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.8 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.4 | GO:0002455 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.1 | GO:0032185 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.1 | GO:0035188 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.5 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.1 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.1 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 0.0 | 0.1 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.1 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.1 | GO:0001990 | regulation of systemic arterial blood pressure by hormone(GO:0001990) |
| 0.0 | 0.1 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) positive regulation of vasoconstriction(GO:0045907) |
| 0.0 | 0.0 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.5 | 1.8 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.4 | 2.2 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.3 | 1.0 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.2 | 1.3 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.2 | 0.5 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) anchored component of the cytoplasmic side of the plasma membrane(GO:0098753) |
| 0.2 | 1.8 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 0.7 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 1.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 8.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 1.4 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.5 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 0.3 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.1 | 0.6 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 0.5 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.1 | 0.4 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.1 | 0.3 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 0.4 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 0.4 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.1 | 2.9 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 5.3 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.1 | 0.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 3.9 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 0.3 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 0.7 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.1 | 0.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.3 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.5 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.7 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 1.9 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 1.0 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 1.5 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.3 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 2.6 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.9 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.2 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.0 | 0.3 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.6 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.0 | 0.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 1.4 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 1.1 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.5 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.3 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 1.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.2 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.5 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 1.1 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 1.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.6 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 2.9 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 1.4 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.9 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.6 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.2 | GO:0098835 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.0 | 0.1 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 2.1 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 1.4 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.2 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.0 | 1.0 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 2.3 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.6 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.8 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0005921 | gap junction(GO:0005921) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.4 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.4 | 1.3 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.4 | 5.8 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.3 | 1.0 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.3 | 0.8 | GO:0005335 | serotonin:sodium symporter activity(GO:0005335) serotonin transmembrane transporter activity(GO:0015222) |
| 0.2 | 0.7 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) cholesterol 26-hydroxylase activity(GO:0031073) |
| 0.2 | 0.9 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.2 | 3.4 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.2 | 0.9 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.2 | 1.1 | GO:0051430 | mu-type opioid receptor binding(GO:0031852) dopamine receptor binding(GO:0050780) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.2 | 0.6 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.2 | 1.0 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.2 | 1.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.2 | 0.9 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.2 | 1.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.2 | 1.0 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.2 | 0.6 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.1 | 0.6 | GO:0015432 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.1 | 0.4 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.1 | 0.7 | GO:0090556 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.1 | 0.5 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.1 | 0.4 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.1 | 10.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.4 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.1 | 1.4 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 0.7 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 0.9 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.3 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.1 | 2.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.4 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 0.8 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.1 | 0.6 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 0.3 | GO:0052834 | inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 0.8 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.1 | 0.8 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.1 | 0.5 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.1 | 0.8 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 0.3 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.1 | 0.5 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 0.9 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.3 | GO:0010852 | cyclase inhibitor activity(GO:0010852) guanylate cyclase inhibitor activity(GO:0030251) |
| 0.1 | 0.3 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.1 | 0.5 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.1 | 0.4 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.1 | 0.3 | GO:0080132 | fatty acid alpha-hydroxylase activity(GO:0080132) |
| 0.1 | 0.5 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.1 | 2.5 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.3 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.1 | 0.3 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 0.8 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 3.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.2 | GO:0003999 | adenine phosphoribosyltransferase activity(GO:0003999) |
| 0.1 | 0.6 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.1 | 0.2 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 0.3 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.1 | 0.6 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.1 | 0.7 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.3 | GO:0015182 | L-histidine transmembrane transporter activity(GO:0005290) L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.1 | 0.7 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 1.1 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 0.8 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.1 | 1.5 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 0.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.6 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 1.5 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 1.6 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 0.2 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.1 | 0.3 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 0.5 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.1 | 0.5 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.3 | GO:1990931 | DNA-N1-methyladenine dioxygenase activity(GO:0043734) RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.1 | 0.4 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 2.0 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 0.2 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.1 | 0.2 | GO:0030791 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.1 | 1.0 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 0.8 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.1 | 0.5 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 0.8 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.4 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 1.1 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 0.2 | GO:0016713 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.1 | 0.9 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 2.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 1.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.6 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.4 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 0.3 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 0.5 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.5 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.4 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.1 | 0.2 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 0.3 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.1 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 0.0 | 0.2 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.2 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 3.1 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.4 | GO:0015154 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.3 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.2 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.0 | 0.4 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.6 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.2 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.1 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.2 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.1 | GO:0033558 | protein deacetylase activity(GO:0033558) NAD-dependent protein deacetylase activity(GO:0034979) |
| 0.0 | 0.5 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.6 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.5 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.1 | GO:0071077 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.0 | 0.3 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) |
| 0.0 | 0.4 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.2 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.2 | GO:0002058 | nucleobase binding(GO:0002054) uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.0 | 1.9 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.2 | GO:0017020 | myosin phosphatase regulator activity(GO:0017020) |
| 0.0 | 0.2 | GO:0010858 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 1.3 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 1.8 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.0 | 0.9 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 1.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.0 | 1.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.7 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.1 | GO:0005183 | gonadotropin hormone-releasing hormone activity(GO:0005183) gonadotropin-releasing hormone receptor binding(GO:0031530) |
| 0.0 | 0.2 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.0 | 4.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.2 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.2 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.3 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.1 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.0 | 1.0 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.4 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.9 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.2 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.0 | 0.4 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.5 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.7 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.2 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 0.1 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.8 | GO:0009975 | cyclase activity(GO:0009975) |
| 0.0 | 0.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 1.8 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.4 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.2 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.5 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.2 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.3 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 5.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 3.9 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.5 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.2 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.2 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.2 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.1 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.1 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.3 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.1 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.2 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.2 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.8 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.0 | 0.8 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.2 | GO:0030546 | receptor activator activity(GO:0030546) receptor agonist activity(GO:0048018) |
| 0.0 | 1.1 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 1.5 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.3 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.1 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 0.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.2 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.4 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.0 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.4 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.6 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.1 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.1 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 2.9 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.3 | GO:0036442 | hydrogen-exporting ATPase activity(GO:0036442) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.1 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 0.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.2 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.2 | GO:0005539 | glycosaminoglycan binding(GO:0005539) |
| 0.0 | 0.0 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 0.5 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 2.1 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 3.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 1.7 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.1 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 1.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.2 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.9 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.3 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.5 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.3 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 2.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.2 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.1 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.2 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.1 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.2 | 0.9 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.2 | 2.6 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.1 | 3.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 0.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 0.6 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 1.3 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.1 | 1.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 0.5 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 4.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 3.3 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 1.3 | REACTOME GABA RECEPTOR ACTIVATION | Genes involved in GABA receptor activation |
| 0.1 | 0.7 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 0.9 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 0.4 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 0.2 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.1 | 0.9 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 1.1 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.3 | REACTOME AQUAPORIN MEDIATED TRANSPORT | Genes involved in Aquaporin-mediated transport |
| 0.0 | 0.7 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.7 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.6 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.3 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.6 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.5 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.3 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.6 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.7 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.4 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.1 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.1 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.1 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.3 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.3 | REACTOME APOPTOTIC EXECUTION PHASE | Genes involved in Apoptotic execution phase |
| 0.0 | 0.2 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.4 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |