Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
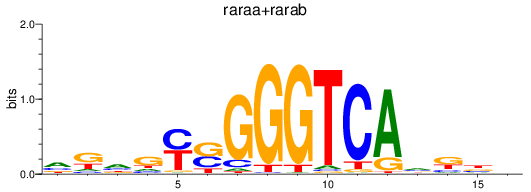
Results for raraa+rarab
Z-value: 0.18
Transcription factors associated with raraa+rarab
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rarab
|
ENSDARG00000034893 | retinoic acid receptor, alpha b |
|
raraa
|
ENSDARG00000056783 | retinoic acid receptor, alpha a |
|
rarab
|
ENSDARG00000111757 | retinoic acid receptor, alpha b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rarab | dr11_v1_chr3_-_33175583_33175583 | 0.31 | 2.0e-01 | Click! |
| raraa | dr11_v1_chr12_+_11007286_11007286 | -0.13 | 5.9e-01 | Click! |
Activity profile of raraa+rarab motif
Sorted Z-values of raraa+rarab motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_28445052 | 0.17 |
ENSDART00000077871
|
pygma
|
phosphorylase, glycogen, muscle A |
| chr17_+_33340675 | 0.13 |
ENSDART00000184396
ENSDART00000077553 |
xdh
|
xanthine dehydrogenase |
| chr14_-_50892442 | 0.13 |
ENSDART00000174562
|
CDHR2
|
cadherin related family member 2 |
| chr3_+_33367954 | 0.11 |
ENSDART00000103161
|
mpp2a
|
membrane protein, palmitoylated 2a (MAGUK p55 subfamily member 2) |
| chr2_+_52847049 | 0.10 |
ENSDART00000121980
|
creb3l3b
|
cAMP responsive element binding protein 3-like 3b |
| chr8_-_28342313 | 0.09 |
ENSDART00000005583
|
kdm5bb
|
lysine (K)-specific demethylase 5Bb |
| chr6_+_103361 | 0.08 |
ENSDART00000151899
|
ldlrb
|
low density lipoprotein receptor b |
| chr1_+_57348756 | 0.08 |
ENSDART00000063750
|
b3gntl1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase-like 1 |
| chr15_+_1199407 | 0.08 |
ENSDART00000163827
|
mfsd1
|
major facilitator superfamily domain containing 1 |
| chr17_+_14711765 | 0.07 |
ENSDART00000012889
|
cx28.6
|
connexin 28.6 |
| chr14_-_2001386 | 0.07 |
ENSDART00000170346
ENSDART00000163975 |
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr2_-_16449504 | 0.07 |
ENSDART00000144801
|
atr
|
ATR serine/threonine kinase |
| chr2_+_58841181 | 0.07 |
ENSDART00000164102
|
cirbpa
|
cold inducible RNA binding protein a |
| chr2_+_56213694 | 0.06 |
ENSDART00000162582
|
upf1
|
upf1 regulator of nonsense transcripts homolog (yeast) |
| chr18_+_3338228 | 0.05 |
ENSDART00000161520
|
gdpd4a
|
glycerophosphodiester phosphodiesterase domain containing 4a |
| chr17_+_53311618 | 0.05 |
ENSDART00000166517
|
asb2b
|
ankyrin repeat and SOCS box containing 2b |
| chr4_-_191736 | 0.05 |
ENSDART00000169187
ENSDART00000192054 |
ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr15_+_47582207 | 0.05 |
ENSDART00000159388
|
CABZ01087566.1
|
|
| chr14_+_94946 | 0.05 |
ENSDART00000165766
ENSDART00000163778 |
mcm7
|
minichromosome maintenance complex component 7 |
| chr14_+_94603 | 0.05 |
ENSDART00000162480
|
mcm7
|
minichromosome maintenance complex component 7 |
| chr14_-_51855047 | 0.05 |
ENSDART00000088912
|
cplx1
|
complexin 1 |
| chr6_-_8277221 | 0.05 |
ENSDART00000053869
|
slc44a2
|
solute carrier family 44 (choline transporter), member 2 |
| chr24_-_36727922 | 0.04 |
ENSDART00000135142
|
si:ch73-334d15.1
|
si:ch73-334d15.1 |
| chr18_+_43183749 | 0.04 |
ENSDART00000151166
|
nectin1b
|
nectin cell adhesion molecule 1b |
| chr18_+_7073130 | 0.04 |
ENSDART00000101216
ENSDART00000148947 |
si:dkey-88e18.2
|
si:dkey-88e18.2 |
| chr19_+_19762183 | 0.04 |
ENSDART00000163611
ENSDART00000187604 |
hoxa3a
|
homeobox A3a |
| chr23_-_21758253 | 0.04 |
ENSDART00000046613
|
vps13d
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr8_+_37168570 | 0.04 |
ENSDART00000098636
|
csf1b
|
colony stimulating factor 1b (macrophage) |
| chr2_+_3696038 | 0.04 |
ENSDART00000150499
ENSDART00000136906 ENSDART00000164152 ENSDART00000128514 ENSDART00000108964 |
egfra
|
epidermal growth factor receptor a (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) |
| chr3_+_57268363 | 0.04 |
ENSDART00000180725
|
TMEM235 (1 of many)
|
transmembrane protein 235 |
| chr21_-_43482426 | 0.04 |
ENSDART00000192901
|
ankrd46a
|
ankyrin repeat domain 46a |
| chr5_-_3885727 | 0.04 |
ENSDART00000143250
|
mlxipl
|
MLX interacting protein like |
| chr22_+_21324398 | 0.04 |
ENSDART00000168509
|
shc2
|
SHC (Src homology 2 domain containing) transforming protein 2 |
| chr25_-_3132060 | 0.03 |
ENSDART00000014363
|
epx
|
eosinophil peroxidase |
| chr24_+_37723362 | 0.03 |
ENSDART00000136836
|
rab11fip3
|
RAB11 family interacting protein 3 (class II) |
| chr23_-_3703569 | 0.02 |
ENSDART00000143731
|
pacsin1a
|
protein kinase C and casein kinase substrate in neurons 1a |
| chr13_+_28618086 | 0.02 |
ENSDART00000087001
|
cnnm2a
|
cyclin and CBS domain divalent metal cation transport mediator 2a |
| chr9_+_500052 | 0.02 |
ENSDART00000166707
|
CU984600.1
|
|
| chr8_-_13454281 | 0.02 |
ENSDART00000141959
|
CR354547.2
|
|
| chr22_+_30009926 | 0.02 |
ENSDART00000142529
|
si:dkey-286j15.1
|
si:dkey-286j15.1 |
| chr4_+_1530287 | 0.02 |
ENSDART00000067446
|
slc38a4
|
solute carrier family 38, member 4 |
| chr24_+_42117231 | 0.02 |
ENSDART00000067734
|
snx16
|
sorting nexin 16 |
| chr21_+_7332963 | 0.02 |
ENSDART00000169834
|
AP3B1
|
adaptor related protein complex 3 beta 1 subunit |
| chr21_+_2506013 | 0.02 |
ENSDART00000162351
|
hmgcrb
|
3-hydroxy-3-methylglutaryl-CoA reductase b |
| chr8_-_13442871 | 0.02 |
ENSDART00000144887
|
pimr106
|
Pim proto-oncogene, serine/threonine kinase, related 106 |
| chr11_+_1608348 | 0.01 |
ENSDART00000162438
|
si:dkey-40c23.3
|
si:dkey-40c23.3 |
| chr19_+_7938121 | 0.01 |
ENSDART00000140053
ENSDART00000146649 |
si:dkey-266f7.5
|
si:dkey-266f7.5 |
| chr12_+_2677303 | 0.01 |
ENSDART00000093113
|
antxr1c
|
anthrax toxin receptor 1c |
| chr10_-_41450367 | 0.01 |
ENSDART00000122682
ENSDART00000189549 |
cabp1b
|
calcium binding protein 1b |
| chr22_+_20208185 | 0.01 |
ENSDART00000142748
|
si:dkey-110c1.7
|
si:dkey-110c1.7 |
| chr18_+_1145571 | 0.01 |
ENSDART00000135055
|
rec114
|
REC114 meiotic recombination protein |
| chr17_+_14886828 | 0.01 |
ENSDART00000010507
ENSDART00000131052 |
ptger2a
|
prostaglandin E receptor 2a (subtype EP2) |
| chr18_+_19842569 | 0.01 |
ENSDART00000147470
|
iqch
|
IQ motif containing H |
| chr1_+_58332000 | 0.01 |
ENSDART00000145234
|
ggt1l2.1
|
gamma-glutamyltransferase 1 like 2.1 |
| chr25_+_2263857 | 0.01 |
ENSDART00000076439
|
yars2
|
tyrosyl-tRNA synthetase 2, mitochondrial |
| chr14_-_52552875 | 0.01 |
ENSDART00000170107
|
ppp2r2ba
|
protein phosphatase 2, regulatory subunit B, beta a |
| chr23_+_25305431 | 0.01 |
ENSDART00000143291
|
RPL41
|
si:dkey-151g10.6 |
| chr12_-_2869565 | 0.01 |
ENSDART00000152772
|
zdhhc16b
|
zinc finger, DHHC-type containing 16b |
| chr18_+_10820430 | 0.00 |
ENSDART00000161990
|
mical3a
|
microtubule associated monooxygenase, calponin and LIM domain containing 3a |
| chr8_-_13486258 | 0.00 |
ENSDART00000137459
|
pimr104
|
Pim proto-oncogene, serine/threonine kinase, related 104 |
| chr14_+_36497250 | 0.00 |
ENSDART00000184727
|
TENM3
|
si:dkey-237h12.3 |
| chr8_-_13419049 | 0.00 |
ENSDART00000133656
|
pimr101
|
Pim proto-oncogene, serine/threonine kinase, related 101 |
Network of associatons between targets according to the STRING database.
First level regulatory network of raraa+rarab
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0052576 | carbohydrate localization(GO:0052575) carbohydrate storage(GO:0052576) |
| 0.0 | 0.1 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.0 | 0.0 | GO:0019062 | virion attachment to host cell(GO:0019062) adhesion of symbiont to host(GO:0044406) adhesion of symbiont to host cell(GO:0044650) |
| 0.0 | 0.1 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.0 | 0.0 | GO:0015871 | choline transport(GO:0015871) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0004645 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.0 | 0.1 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.0 | 0.0 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.0 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |