Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
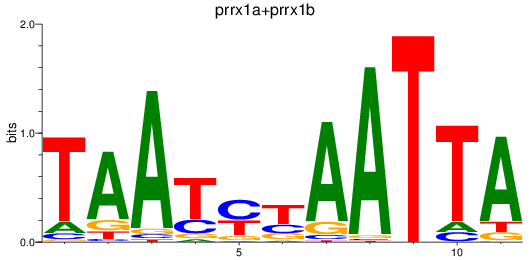
Results for prrx1a+prrx1b
Z-value: 1.02
Transcription factors associated with prrx1a+prrx1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
prrx1a
|
ENSDARG00000033971 | paired related homeobox 1a |
|
prrx1b
|
ENSDARG00000042027 | paired related homeobox 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| prrx1b | dr11_v1_chr20_+_34511678_34511678 | 0.71 | 6.2e-04 | Click! |
| prrx1a | dr11_v1_chr2_-_23172708_23172708 | 0.21 | 3.8e-01 | Click! |
Activity profile of prrx1a+prrx1b motif
Sorted Z-values of prrx1a+prrx1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_31276842 | 14.83 |
ENSDART00000187238
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr1_+_17676745 | 4.77 |
ENSDART00000030665
|
slc25a4
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 |
| chr9_-_22339582 | 4.14 |
ENSDART00000134805
|
crygm2d1
|
crystallin, gamma M2d1 |
| chr2_+_37227011 | 2.94 |
ENSDART00000126587
ENSDART00000084958 |
samd7
|
sterile alpha motif domain containing 7 |
| chr20_-_43775495 | 2.83 |
ENSDART00000100610
ENSDART00000149001 ENSDART00000148809 ENSDART00000100608 |
matn3a
|
matrilin 3a |
| chr1_-_43905252 | 2.45 |
ENSDART00000135477
ENSDART00000132089 |
si:dkey-22i16.3
|
si:dkey-22i16.3 |
| chr4_+_9669717 | 2.41 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr25_+_29160102 | 2.22 |
ENSDART00000162854
|
pkmb
|
pyruvate kinase M1/2b |
| chr25_+_31277415 | 2.19 |
ENSDART00000036275
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr15_+_45640906 | 2.14 |
ENSDART00000149361
ENSDART00000149079 |
sagb
|
S-antigen; retina and pineal gland (arrestin) b |
| chr11_+_36243774 | 2.11 |
ENSDART00000023323
|
zgc:172270
|
zgc:172270 |
| chr23_-_24343363 | 2.00 |
ENSDART00000166392
|
fam131c
|
family with sequence similarity 131, member C |
| chr15_-_44512461 | 1.96 |
ENSDART00000155456
|
gria4a
|
glutamate receptor, ionotropic, AMPA 4a |
| chr25_+_31227747 | 1.95 |
ENSDART00000033872
|
tnni2a.1
|
troponin I type 2a (skeletal, fast), tandem duplicate 1 |
| chr24_-_7697274 | 1.74 |
ENSDART00000186077
|
syt5b
|
synaptotagmin Vb |
| chr7_+_35075847 | 1.70 |
ENSDART00000193469
ENSDART00000037346 |
ctrb1
|
chymotrypsinogen B1 |
| chr21_+_7582036 | 1.70 |
ENSDART00000135485
ENSDART00000027268 |
otpa
|
orthopedia homeobox a |
| chr14_+_11430796 | 1.68 |
ENSDART00000165275
|
si:ch211-153b23.3
|
si:ch211-153b23.3 |
| chr17_-_19345521 | 1.66 |
ENSDART00000082085
|
gsc
|
goosecoid |
| chr7_+_66822229 | 1.64 |
ENSDART00000112109
|
lyve1a
|
lymphatic vessel endothelial hyaluronic receptor 1a |
| chr4_-_14315855 | 1.62 |
ENSDART00000133325
|
nell2b
|
neural EGFL like 2b |
| chr2_+_38039857 | 1.61 |
ENSDART00000159951
|
casq1a
|
calsequestrin 1a |
| chr14_-_49063157 | 1.58 |
ENSDART00000021260
|
sept8b
|
septin 8b |
| chr16_-_45058919 | 1.58 |
ENSDART00000177134
|
gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr3_-_31079186 | 1.57 |
ENSDART00000145636
ENSDART00000140569 |
ELOB (1 of many)
elob
|
elongin B elongin B |
| chr21_-_27881752 | 1.53 |
ENSDART00000132583
|
nrxn2a
|
neurexin 2a |
| chr6_+_40661703 | 1.53 |
ENSDART00000142492
|
eno1b
|
enolase 1b, (alpha) |
| chr6_+_40354424 | 1.46 |
ENSDART00000047416
|
slc4a8
|
solute carrier family 4, sodium bicarbonate cotransporter, member 8 |
| chr15_-_16098531 | 1.46 |
ENSDART00000080377
|
aldoca
|
aldolase C, fructose-bisphosphate, a |
| chr9_+_34641237 | 1.44 |
ENSDART00000133996
|
shox
|
short stature homeobox |
| chr22_-_910926 | 1.41 |
ENSDART00000180075
|
FP016205.1
|
|
| chr13_+_25449681 | 1.38 |
ENSDART00000101328
|
atoh7
|
atonal bHLH transcription factor 7 |
| chr7_-_28148310 | 1.38 |
ENSDART00000044208
|
lmo1
|
LIM domain only 1 |
| chr3_-_32337653 | 1.33 |
ENSDART00000156918
ENSDART00000156551 |
si:dkey-16p21.8
|
si:dkey-16p21.8 |
| chr7_-_13884610 | 1.31 |
ENSDART00000006897
|
rlbp1a
|
retinaldehyde binding protein 1a |
| chr17_-_16965809 | 1.29 |
ENSDART00000153697
|
nrxn3a
|
neurexin 3a |
| chr15_+_19797918 | 1.28 |
ENSDART00000113314
|
si:ch211-229d2.5
|
si:ch211-229d2.5 |
| chr22_+_18389271 | 1.26 |
ENSDART00000088270
|
yjefn3
|
YjeF N-terminal domain containing 3 |
| chr10_+_29698467 | 1.25 |
ENSDART00000163402
|
dlg2
|
discs, large homolog 2 (Drosophila) |
| chr23_+_45584223 | 1.25 |
ENSDART00000149367
|
si:ch73-290k24.5
|
si:ch73-290k24.5 |
| chr11_+_27543093 | 1.24 |
ENSDART00000023889
|
barx1
|
BARX homeobox 1 |
| chr10_-_26744131 | 1.23 |
ENSDART00000020096
ENSDART00000162710 ENSDART00000179853 |
fgf13b
|
fibroblast growth factor 13b |
| chr16_-_27749172 | 1.22 |
ENSDART00000145198
|
steap4
|
STEAP family member 4 |
| chr19_+_9174166 | 1.22 |
ENSDART00000104637
ENSDART00000150968 |
si:ch211-81a5.8
|
si:ch211-81a5.8 |
| chr19_+_10339538 | 1.21 |
ENSDART00000151808
ENSDART00000151235 |
rcvrn3
|
recoverin 3 |
| chr7_-_27685365 | 1.19 |
ENSDART00000188342
|
calca
|
calcitonin/calcitonin-related polypeptide, alpha |
| chr18_-_16801033 | 1.19 |
ENSDART00000100100
|
admb
|
adrenomedullin b |
| chr1_+_37391141 | 1.18 |
ENSDART00000083593
ENSDART00000168647 |
sparcl1
|
SPARC-like 1 |
| chr21_+_27382893 | 1.13 |
ENSDART00000005682
|
actn3a
|
actinin alpha 3a |
| chr20_-_9462433 | 1.13 |
ENSDART00000152674
ENSDART00000040557 |
zgc:101840
|
zgc:101840 |
| chr1_-_50859053 | 1.10 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr3_-_32170850 | 1.06 |
ENSDART00000055307
ENSDART00000157366 |
tnnt1
|
troponin T type 1 (skeletal, slow) |
| chr20_-_44496245 | 1.06 |
ENSDART00000012229
|
fkbp1b
|
FK506 binding protein 1b |
| chr9_-_46701326 | 1.05 |
ENSDART00000159971
|
si:ch73-193i22.1
|
si:ch73-193i22.1 |
| chr25_-_210730 | 1.05 |
ENSDART00000187580
|
FP236318.1
|
|
| chr21_+_19008168 | 1.04 |
ENSDART00000136196
ENSDART00000128381 ENSDART00000176624 |
nefla
|
neurofilament, light polypeptide a |
| chr3_-_24980067 | 1.03 |
ENSDART00000048871
|
desi1a
|
desumoylating isopeptidase 1a |
| chr25_+_26921480 | 1.00 |
ENSDART00000155949
|
grm8b
|
glutamate receptor, metabotropic 8b |
| chr20_-_2134620 | 0.99 |
ENSDART00000064375
|
tmem244
|
transmembrane protein 244 |
| chr11_-_37509001 | 0.99 |
ENSDART00000109753
|
bsnb
|
bassoon (presynaptic cytomatrix protein) b |
| chr21_+_26720803 | 0.98 |
ENSDART00000053797
|
slc3a2b
|
solute carrier family 3 (amino acid transporter heavy chain), member 2b |
| chr1_+_25801648 | 0.98 |
ENSDART00000129471
|
gucy1b1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr13_-_29421331 | 0.97 |
ENSDART00000150228
|
chata
|
choline O-acetyltransferase a |
| chr18_+_8833251 | 0.95 |
ENSDART00000143519
|
impdh1a
|
IMP (inosine 5'-monophosphate) dehydrogenase 1a |
| chr18_+_1703984 | 0.95 |
ENSDART00000114010
|
slitrk3a
|
SLIT and NTRK-like family, member 3a |
| chr3_+_39540014 | 0.95 |
ENSDART00000074848
|
zgc:165423
|
zgc:165423 |
| chr3_+_13440900 | 0.93 |
ENSDART00000143715
|
GNG14
|
si:dkey-117i10.1 |
| chr10_-_10607118 | 0.93 |
ENSDART00000101089
|
dbh
|
dopamine beta-hydroxylase (dopamine beta-monooxygenase) |
| chr5_+_11407504 | 0.91 |
ENSDART00000186723
ENSDART00000018047 |
zgc:112294
|
zgc:112294 |
| chr15_-_21702317 | 0.91 |
ENSDART00000155824
|
IL4I1
|
si:dkey-40g16.6 |
| chr17_-_37214196 | 0.90 |
ENSDART00000128715
|
kif3cb
|
kinesin family member 3Cb |
| chr23_-_12345764 | 0.90 |
ENSDART00000133956
|
phactr3a
|
phosphatase and actin regulator 3a |
| chr10_-_34772211 | 0.90 |
ENSDART00000145450
ENSDART00000134307 |
dclk1a
|
doublecortin-like kinase 1a |
| chr25_-_13381854 | 0.89 |
ENSDART00000164621
ENSDART00000169129 |
ndrg4
|
NDRG family member 4 |
| chr18_-_47662696 | 0.87 |
ENSDART00000184260
|
CABZ01073963.1
|
|
| chr25_+_31267268 | 0.87 |
ENSDART00000181239
|
tnni2a.3
|
troponin I type 2a (skeletal, fast), tandem duplicate 3 |
| chr21_-_43022048 | 0.86 |
ENSDART00000138329
|
dpysl3
|
dihydropyrimidinase-like 3 |
| chr21_+_3093419 | 0.85 |
ENSDART00000162520
|
SHC3
|
SHC adaptor protein 3 |
| chr17_+_18117029 | 0.85 |
ENSDART00000154646
ENSDART00000179739 |
bcl11ba
|
B cell CLL/lymphoma 11Ba |
| chr1_-_10647484 | 0.84 |
ENSDART00000164541
ENSDART00000188958 ENSDART00000190904 |
si:dkey-31e10.1
|
si:dkey-31e10.1 |
| chr21_+_6556635 | 0.83 |
ENSDART00000139598
|
col5a1
|
procollagen, type V, alpha 1 |
| chr25_+_20089986 | 0.83 |
ENSDART00000143441
ENSDART00000184073 |
tnni4b.2
|
troponin I4b, tandem duplicate 2 |
| chr18_-_8885792 | 0.82 |
ENSDART00000143619
|
si:dkey-95h12.1
|
si:dkey-95h12.1 |
| chr12_-_20795867 | 0.82 |
ENSDART00000152835
ENSDART00000153424 |
nme2a
|
NME/NM23 nucleoside diphosphate kinase 2a |
| chr7_+_25858380 | 0.82 |
ENSDART00000148780
ENSDART00000079218 |
mtmr1a
|
myotubularin related protein 1a |
| chr12_+_42436920 | 0.82 |
ENSDART00000177303
|
ebf3a
|
early B cell factor 3a |
| chr21_+_13861589 | 0.81 |
ENSDART00000015629
ENSDART00000171306 |
stxbp1a
|
syntaxin binding protein 1a |
| chr12_+_18681477 | 0.81 |
ENSDART00000127981
ENSDART00000143979 |
rgs9b
|
regulator of G protein signaling 9b |
| chr19_-_5103313 | 0.81 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr4_+_9279784 | 0.81 |
ENSDART00000014897
|
srgap1b
|
SLIT-ROBO Rho GTPase activating protein 1b |
| chr14_+_22152060 | 0.80 |
ENSDART00000082697
ENSDART00000054982 |
gabra6a
|
gamma-aminobutyric acid (GABA) A receptor, alpha 6a |
| chr4_-_390431 | 0.79 |
ENSDART00000067482
ENSDART00000138500 |
dynlt1
|
dynein, light chain, Tctex-type 1 |
| chr14_+_19258702 | 0.78 |
ENSDART00000187087
ENSDART00000005738 |
slitrk2
|
SLIT and NTRK-like family, member 2 |
| chr20_-_30920356 | 0.78 |
ENSDART00000022951
|
kif25
|
kinesin family member 25 |
| chr6_-_40884453 | 0.78 |
ENSDART00000017968
ENSDART00000154100 |
sirt4
|
sirtuin 4 |
| chr12_+_42436328 | 0.77 |
ENSDART00000167324
|
ebf3a
|
early B cell factor 3a |
| chr7_-_35515931 | 0.76 |
ENSDART00000193324
|
irx6a
|
iroquois homeobox 6a |
| chr22_+_20208185 | 0.76 |
ENSDART00000142748
|
si:dkey-110c1.7
|
si:dkey-110c1.7 |
| chr10_-_34741738 | 0.75 |
ENSDART00000163072
|
dclk1a
|
doublecortin-like kinase 1a |
| chr16_-_16643377 | 0.75 |
ENSDART00000185748
|
tnxba
|
tenascin XBa |
| chr4_+_2267641 | 0.75 |
ENSDART00000165503
|
si:ch73-89b15.3
|
si:ch73-89b15.3 |
| chr7_+_26629084 | 0.74 |
ENSDART00000101044
ENSDART00000173765 |
hsbp1a
|
heat shock factor binding protein 1a |
| chr13_+_24279021 | 0.72 |
ENSDART00000058629
|
acta1b
|
actin, alpha 1b, skeletal muscle |
| chr8_+_2575993 | 0.72 |
ENSDART00000112914
|
si:ch211-51h9.7
|
si:ch211-51h9.7 |
| chr8_-_18203274 | 0.71 |
ENSDART00000134078
ENSDART00000180235 ENSDART00000080006 ENSDART00000125418 ENSDART00000142114 |
elovl8b
|
ELOVL fatty acid elongase 8b |
| chr19_+_31771270 | 0.71 |
ENSDART00000147474
|
stmn2b
|
stathmin 2b |
| chr6_+_37894220 | 0.70 |
ENSDART00000087311
|
oca2
|
oculocutaneous albinism II |
| chr15_-_576135 | 0.69 |
ENSDART00000124170
|
cbln20
|
cerebellin 20 |
| chr13_+_31757331 | 0.69 |
ENSDART00000044282
|
hif1aa
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) a |
| chr9_-_48397702 | 0.68 |
ENSDART00000147169
|
zgc:172182
|
zgc:172182 |
| chr9_+_35876927 | 0.68 |
ENSDART00000138834
|
mab21l3
|
mab-21-like 3 |
| chr25_+_3327071 | 0.68 |
ENSDART00000136131
ENSDART00000133243 |
ldhbb
|
lactate dehydrogenase Bb |
| chr12_-_47601845 | 0.67 |
ENSDART00000169548
ENSDART00000182889 |
rgs7b
|
regulator of G protein signaling 7b |
| chr16_-_41439659 | 0.67 |
ENSDART00000191624
|
cpne4a
|
copine IVa |
| chr5_-_69180587 | 0.67 |
ENSDART00000156681
ENSDART00000160753 |
zgc:171967
|
zgc:171967 |
| chr19_-_5103141 | 0.67 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr20_+_34717403 | 0.66 |
ENSDART00000034252
|
pnocb
|
prepronociceptin b |
| chr5_+_36850650 | 0.65 |
ENSDART00000051186
|
nccrp1
|
non-specific cytotoxic cell receptor protein 1 |
| chr24_-_38384432 | 0.65 |
ENSDART00000140739
|
lrrc4bb
|
leucine rich repeat containing 4Bb |
| chr14_-_34044369 | 0.64 |
ENSDART00000149396
ENSDART00000123607 ENSDART00000190746 |
cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr2_+_23823622 | 0.63 |
ENSDART00000099581
|
si:dkey-24c2.9
|
si:dkey-24c2.9 |
| chr16_+_50434668 | 0.62 |
ENSDART00000193500
|
IGLON5
|
zgc:110372 |
| chr6_+_52350443 | 0.62 |
ENSDART00000151612
ENSDART00000151349 |
si:ch211-239j9.1
|
si:ch211-239j9.1 |
| chr23_+_34047413 | 0.61 |
ENSDART00000143933
ENSDART00000123925 ENSDART00000176139 |
chchd6a
|
coiled-coil-helix-coiled-coil-helix domain containing 6a |
| chr2_-_5505094 | 0.61 |
ENSDART00000145035
|
saga
|
S-antigen; retina and pineal gland (arrestin) a |
| chr22_-_15562933 | 0.61 |
ENSDART00000141528
|
ankmy1
|
ankyrin repeat and MYND domain containing 1 |
| chr24_-_38012593 | 0.60 |
ENSDART00000151970
|
si:ch211-248l17.3
|
si:ch211-248l17.3 |
| chr25_+_22730490 | 0.60 |
ENSDART00000149455
|
abcc8
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 8 |
| chr18_-_26785861 | 0.60 |
ENSDART00000098361
|
nmba
|
neuromedin Ba |
| chr23_-_31763753 | 0.59 |
ENSDART00000053399
|
aldh8a1
|
aldehyde dehydrogenase 8 family, member A1 |
| chr2_+_56463167 | 0.59 |
ENSDART00000123392
|
rab11bb
|
RAB11B, member RAS oncogene family, b |
| chr3_-_55404985 | 0.59 |
ENSDART00000154274
|
mafga
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Ga |
| chr3_+_28953274 | 0.59 |
ENSDART00000133528
ENSDART00000103602 |
lgals2a
|
lectin, galactoside-binding, soluble, 2a |
| chr1_-_45553602 | 0.59 |
ENSDART00000143664
|
grin2bb
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2B, genome duplicate b |
| chr13_-_49444636 | 0.58 |
ENSDART00000136991
|
irf2bp2a
|
interferon regulatory factor 2 binding protein 2a |
| chr8_-_44868020 | 0.58 |
ENSDART00000142712
|
cacna1fa
|
calcium channel, voltage-dependent, L type, alpha 1F subunit a |
| chr10_-_33621739 | 0.58 |
ENSDART00000142655
ENSDART00000128049 |
hunk
|
hormonally up-regulated Neu-associated kinase |
| chr1_+_12009673 | 0.58 |
ENSDART00000080100
|
slc24a2
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 2 |
| chr22_-_10121880 | 0.58 |
ENSDART00000002348
|
rdh5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr17_+_46387086 | 0.57 |
ENSDART00000157079
|
si:dkey-206p8.1
|
si:dkey-206p8.1 |
| chr7_+_20344032 | 0.57 |
ENSDART00000144948
ENSDART00000138786 |
ponzr1
|
plac8 onzin related protein 1 |
| chr18_+_38321039 | 0.56 |
ENSDART00000132534
ENSDART00000111260 ENSDART00000192806 |
alx4b
|
ALX homeobox 4b |
| chr10_-_17083180 | 0.55 |
ENSDART00000170083
|
fam166b
|
family with sequence similarity 166, member B |
| chr8_+_635704 | 0.55 |
ENSDART00000130358
|
csgalnact1b
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1b |
| chr14_-_17068712 | 0.54 |
ENSDART00000170277
|
phox2bb
|
paired-like homeobox 2bb |
| chr17_-_15657029 | 0.54 |
ENSDART00000153925
|
fut9a
|
fucosyltransferase 9a |
| chr12_-_558201 | 0.54 |
ENSDART00000168586
ENSDART00000158355 |
bsk146
|
brain specific kinase 146 |
| chr25_-_19648154 | 0.53 |
ENSDART00000148570
|
atp2b1b
|
ATPase plasma membrane Ca2+ transporting 1b |
| chr12_+_17504559 | 0.53 |
ENSDART00000020628
|
cyth3a
|
cytohesin 3a |
| chr2_+_15203322 | 0.53 |
ENSDART00000144171
|
abca4b
|
ATP-binding cassette, sub-family A (ABC1), member 4b |
| chr6_+_8315050 | 0.53 |
ENSDART00000189987
|
gcdha
|
glutaryl-CoA dehydrogenase a |
| chr18_+_5490668 | 0.52 |
ENSDART00000167035
|
mibp2
|
muscle-specific beta 1 integrin binding protein 2 |
| chr12_-_25294769 | 0.52 |
ENSDART00000153306
|
hcar1-4
|
hydroxycarboxylic acid receptor 1-4 |
| chr24_+_25069609 | 0.52 |
ENSDART00000115165
|
amer2
|
APC membrane recruitment protein 2 |
| chr8_+_21406769 | 0.51 |
ENSDART00000135766
|
si:dkey-163f12.6
|
si:dkey-163f12.6 |
| chr12_-_19091214 | 0.51 |
ENSDART00000153225
|
si:ch73-139e5.4
|
si:ch73-139e5.4 |
| chr22_+_1556948 | 0.50 |
ENSDART00000159050
|
si:ch211-255f4.8
|
si:ch211-255f4.8 |
| chr9_-_48407408 | 0.50 |
ENSDART00000058248
|
zgc:172182
|
zgc:172182 |
| chr4_+_72578191 | 0.50 |
ENSDART00000182434
|
si:cabz01054394.5
|
si:cabz01054394.5 |
| chr13_-_23095006 | 0.49 |
ENSDART00000089242
|
kif1bp
|
kif1 binding protein |
| chr8_+_41647539 | 0.49 |
ENSDART00000136492
ENSDART00000138799 ENSDART00000134404 |
si:ch211-158d24.4
|
si:ch211-158d24.4 |
| chr6_+_23887314 | 0.49 |
ENSDART00000163188
|
znf648
|
zinc finger protein 648 |
| chr25_+_10923100 | 0.48 |
ENSDART00000157055
|
si:ch211-147g22.7
|
si:ch211-147g22.7 |
| chr7_+_20344222 | 0.48 |
ENSDART00000141186
ENSDART00000139274 |
ponzr1
|
plac8 onzin related protein 1 |
| chr24_-_25442342 | 0.48 |
ENSDART00000177140
ENSDART00000138215 |
phex
|
phosphate regulating endopeptidase homolog, X-linked |
| chr22_+_10698549 | 0.48 |
ENSDART00000081228
|
abhd14a
|
abhydrolase domain containing 14A |
| chr23_-_44226556 | 0.47 |
ENSDART00000149115
|
zgc:158659
|
zgc:158659 |
| chr16_+_23984755 | 0.47 |
ENSDART00000145328
|
apoc2
|
apolipoprotein C-II |
| chr23_+_28770225 | 0.47 |
ENSDART00000132179
ENSDART00000142273 |
masp2
|
mannan-binding lectin serine peptidase 2 |
| chr13_-_29505604 | 0.46 |
ENSDART00000110005
|
cdhr1a
|
cadherin-related family member 1a |
| chr3_+_34919810 | 0.46 |
ENSDART00000055264
|
ca10b
|
carbonic anhydrase Xb |
| chr16_+_20161805 | 0.45 |
ENSDART00000192146
|
c16h2orf66
|
chromosome 16 C2orf66 homolog |
| chr25_-_16146851 | 0.45 |
ENSDART00000104043
|
dkk3b
|
dickkopf WNT signaling pathway inhibitor 3b |
| chr23_+_42810055 | 0.45 |
ENSDART00000186647
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr1_+_44814322 | 0.45 |
ENSDART00000059227
|
ndufs8a
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8a |
| chr1_-_30979707 | 0.45 |
ENSDART00000008469
|
dlx2b
|
distal-less homeobox 2b |
| chr3_-_13921173 | 0.45 |
ENSDART00000159177
ENSDART00000165174 |
si:dkey-61n16.5
|
si:dkey-61n16.5 |
| chr16_-_27640995 | 0.44 |
ENSDART00000019658
|
nacad
|
NAC alpha domain containing |
| chr3_+_32594357 | 0.44 |
ENSDART00000151639
|
unc119.2
|
unc-119 lipid binding chaperone B homolog 2 |
| chr13_+_12761707 | 0.44 |
ENSDART00000015127
|
zgc:100846
|
zgc:100846 |
| chr5_+_34407763 | 0.44 |
ENSDART00000188849
ENSDART00000145127 |
lamc3
|
laminin, gamma 3 |
| chr14_+_46291022 | 0.44 |
ENSDART00000074099
|
cabp2b
|
calcium binding protein 2b |
| chr22_+_14051894 | 0.44 |
ENSDART00000142548
|
aox6
|
aldehyde oxidase 6 |
| chr2_+_30916188 | 0.44 |
ENSDART00000137012
|
myom1a
|
myomesin 1a (skelemin) |
| chr4_-_16345227 | 0.44 |
ENSDART00000079521
|
kera
|
keratocan |
| chr18_-_21271373 | 0.43 |
ENSDART00000060001
|
pnp6
|
purine nucleoside phosphorylase 6 |
| chr12_+_10631266 | 0.43 |
ENSDART00000161455
|
csf3a
|
colony stimulating factor 3 (granulocyte) a |
| chr24_-_24282427 | 0.43 |
ENSDART00000123299
|
pdha1b
|
pyruvate dehydrogenase E1 alpha 1 subunit b |
| chr6_+_29693492 | 0.43 |
ENSDART00000114172
|
pde6d
|
phosphodiesterase 6D, cGMP-specific, rod, delta |
| chr20_+_5106568 | 0.42 |
ENSDART00000028039
|
cyp46a1.1
|
cytochrome P450, family 46, subfamily A, polypeptide 1, tandem duplicate 1 |
| chr22_-_23668356 | 0.42 |
ENSDART00000167106
ENSDART00000159622 ENSDART00000163228 |
cfh
|
complement factor H |
| chr16_-_42872571 | 0.42 |
ENSDART00000154757
ENSDART00000102345 |
txnipb
|
thioredoxin interacting protein b |
| chr16_-_16590780 | 0.42 |
ENSDART00000059841
|
si:ch211-257p13.3
|
si:ch211-257p13.3 |
| chr1_+_18811679 | 0.42 |
ENSDART00000078610
|
slc25a51a
|
solute carrier family 25, member 51a |
| chr2_+_20605186 | 0.42 |
ENSDART00000128505
|
olfml2bb
|
olfactomedin-like 2Bb |
| chr5_-_69180227 | 0.41 |
ENSDART00000154816
|
zgc:171967
|
zgc:171967 |
| chr5_-_11944361 | 0.41 |
ENSDART00000165446
ENSDART00000167305 |
rnft2
|
ring finger protein, transmembrane 2 |
| chr14_+_46342882 | 0.40 |
ENSDART00000193707
ENSDART00000060577 |
tmem33
|
transmembrane protein 33 |
| chr6_+_49081141 | 0.40 |
ENSDART00000131080
ENSDART00000050000 |
tshba
|
thyroid stimulating hormone, beta subunit, a |
| chr20_-_46534179 | 0.39 |
ENSDART00000060689
|
eif2b2
|
eukaryotic translation initiation factor 2B, subunit 2 beta |
Network of associatons between targets according to the STRING database.
First level regulatory network of prrx1a+prrx1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.8 | GO:0051503 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.5 | 1.6 | GO:0014809 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.4 | 21.7 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.4 | 1.2 | GO:2000648 | positive regulation of stem cell proliferation(GO:2000648) |
| 0.4 | 1.5 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.3 | 1.0 | GO:0008292 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.3 | 1.3 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.3 | 0.9 | GO:0046333 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.3 | 1.2 | GO:0015677 | copper ion import(GO:0015677) |
| 0.3 | 1.7 | GO:0021767 | mammillary body development(GO:0021767) social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.2 | 1.0 | GO:0099543 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.2 | 0.7 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.2 | 2.7 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.2 | 0.9 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.2 | 0.6 | GO:0097053 | L-kynurenine metabolic process(GO:0097052) L-kynurenine catabolic process(GO:0097053) |
| 0.2 | 0.6 | GO:1904377 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) positive regulation of protein localization to plasma membrane(GO:1903078) positive regulation of protein localization to cell periphery(GO:1904377) |
| 0.2 | 1.5 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.2 | 1.6 | GO:0031579 | membrane raft assembly(GO:0001765) membrane raft organization(GO:0031579) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.2 | 1.6 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.1 | 0.8 | GO:0016081 | synaptic vesicle docking(GO:0016081) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.1 | 0.4 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.1 | 1.2 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.1 | 0.3 | GO:0034375 | high-density lipoprotein particle remodeling(GO:0034375) |
| 0.1 | 0.7 | GO:2001270 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.7 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.1 | 5.3 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.1 | 1.0 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.1 | 1.7 | GO:0010717 | regulation of epithelial to mesenchymal transition(GO:0010717) |
| 0.1 | 0.6 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.1 | 1.0 | GO:0072078 | renal tubule morphogenesis(GO:0061333) nephron tubule morphogenesis(GO:0072078) nephron epithelium morphogenesis(GO:0072088) |
| 0.1 | 1.0 | GO:0046037 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.1 | 0.3 | GO:0060879 | peripheral nervous system myelin formation(GO:0032290) semicircular canal fusion(GO:0060879) |
| 0.1 | 1.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.6 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 | 0.3 | GO:0030186 | melatonin metabolic process(GO:0030186) |
| 0.1 | 0.5 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 1.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 1.3 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.5 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.1 | 1.4 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 0.4 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.1 | 0.7 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 0.5 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 0.3 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.1 | 0.9 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 1.5 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.4 | GO:0036268 | swimming(GO:0036268) |
| 0.1 | 0.2 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.8 | GO:0006183 | GTP biosynthetic process(GO:0006183) guanosine-containing compound biosynthetic process(GO:1901070) |
| 0.1 | 1.3 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 0.4 | GO:2001286 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.1 | 0.3 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.1 | 1.1 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.4 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.0 | 0.6 | GO:0046887 | positive regulation of hormone secretion(GO:0046887) |
| 0.0 | 0.1 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 0.0 | 0.2 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.0 | 0.3 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.2 | GO:0032782 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.0 | 0.6 | GO:0001964 | startle response(GO:0001964) |
| 0.0 | 0.7 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.6 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.2 | GO:0071939 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.0 | 1.1 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 1.2 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.4 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.2 | GO:0045046 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.6 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.8 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.0 | 0.4 | GO:0060142 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) |
| 0.0 | 0.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.5 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 1.1 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.5 | GO:0090303 | positive regulation of wound healing(GO:0090303) |
| 0.0 | 0.6 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.0 | 0.2 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 1.4 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.4 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.2 | GO:0034695 | response to prostaglandin E(GO:0034695) cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.0 | 1.6 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 0.5 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 1.2 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.5 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 0.7 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.7 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.0 | 0.2 | GO:0030431 | sleep(GO:0030431) |
| 0.0 | 0.2 | GO:1990709 | presynaptic active zone organization(GO:1990709) |
| 0.0 | 0.4 | GO:0007035 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.0 | 0.1 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.3 | GO:0097300 | necrotic cell death(GO:0070265) programmed necrotic cell death(GO:0097300) |
| 0.0 | 0.5 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.4 | GO:0010257 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 1.4 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 0.8 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.2 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.0 | 0.7 | GO:0050868 | negative regulation of T cell activation(GO:0050868) negative regulation of leukocyte cell-cell adhesion(GO:1903038) |
| 0.0 | 0.8 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.7 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 4.5 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.1 | GO:1902895 | pri-miRNA transcription from RNA polymerase II promoter(GO:0061614) regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902893) positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.0 | 0.4 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.1 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.2 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.0 | 0.8 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.4 | GO:0042159 | lipoprotein catabolic process(GO:0042159) |
| 0.0 | 1.3 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 0.1 | GO:0070285 | pigment cell development(GO:0070285) |
| 0.0 | 0.5 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.3 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.3 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.4 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.0 | 0.2 | GO:0046459 | short-chain fatty acid metabolic process(GO:0046459) |
| 0.0 | 0.7 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.1 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.0 | 0.1 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.0 | 0.1 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 1.4 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.5 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.4 | GO:0060117 | auditory receptor cell development(GO:0060117) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.7 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.1 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.8 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.0 | 0.2 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.2 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.2 | GO:0048016 | calcineurin-NFAT signaling cascade(GO:0033173) inositol phosphate-mediated signaling(GO:0048016) |
| 0.0 | 0.0 | GO:0050968 | chemosensory behavior(GO:0007635) detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.0 | 0.2 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.3 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 0.5 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.7 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.8 | GO:0014904 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.0 | 0.5 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 21.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.3 | 1.6 | GO:0030891 | VCB complex(GO:0030891) |
| 0.2 | 1.6 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.2 | 0.8 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.2 | 1.5 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 1.7 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 2.3 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 1.0 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.1 | 2.0 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 0.4 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.1 | 0.4 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 1.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 0.2 | GO:0008247 | 1-alkyl-2-acetylglycerophosphocholine esterase complex(GO:0008247) |
| 0.1 | 0.9 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.4 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 0.8 | GO:0043679 | axon terminus(GO:0043679) |
| 0.1 | 0.6 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.8 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 0.9 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 3.0 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 0.8 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.5 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.4 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.3 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.7 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 2.0 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.4 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.6 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 1.4 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.4 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.1 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.0 | 1.1 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 1.3 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.2 | GO:0038039 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 1.1 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.4 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 1.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.1 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.0 | 0.2 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.4 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 5.5 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.3 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 1.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.2 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.2 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.3 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.4 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.8 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.2 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.0 | 0.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.8 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.4 | 1.2 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.4 | 1.6 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.4 | 2.2 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.4 | 1.5 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.3 | 0.9 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.3 | 1.2 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.3 | 1.3 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.2 | 1.5 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 1.5 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.2 | 1.0 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.4 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.1 | 1.0 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.1 | 0.6 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.5 | GO:0004361 | glutaryl-CoA dehydrogenase activity(GO:0004361) |
| 0.1 | 2.0 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 0.7 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.1 | 0.6 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.1 | 0.3 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.1 | 1.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.7 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.9 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.5 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 0.4 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.1 | 0.6 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 0.1 | 0.8 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.7 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.5 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.1 | 1.0 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 0.9 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 0.4 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.1 | 1.2 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.1 | 0.2 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.1 | 1.1 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 0.5 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 0.3 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.1 | 0.8 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 1.1 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 1.3 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.1 | 0.7 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 1.0 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 0.8 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 1.1 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.3 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.1 | 0.7 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 4.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 1.1 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 4.6 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 1.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.5 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 1.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.2 | GO:0015126 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.0 | 0.6 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.2 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.8 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 1.7 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 1.0 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.2 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.5 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.3 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.1 | GO:0004776 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.0 | 0.2 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.3 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.1 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.0 | 0.8 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.2 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.0 | 0.3 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.0 | 0.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 1.9 | GO:1902936 | phosphatidylinositol bisphosphate binding(GO:1902936) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 1.1 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.8 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.1 | GO:0015105 | arsenite transmembrane transporter activity(GO:0015105) |
| 0.0 | 0.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.5 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.0 | 0.2 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.2 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.9 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.0 | 0.2 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 0.2 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.1 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.3 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.5 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 1.8 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.2 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.3 | GO:0016917 | GABA receptor activity(GO:0016917) |
| 0.0 | 0.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0052794 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.0 | GO:0072570 | ADP-D-ribose binding(GO:0072570) mono-ADP-D-ribose binding(GO:0072571) |
| 0.0 | 2.3 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 1.2 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 2.3 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.2 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 0.4 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.7 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.3 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.2 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.1 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.2 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.3 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 4.8 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.1 | 0.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 0.9 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 0.9 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 0.8 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 0.8 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 1.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.6 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 1.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.3 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 1.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 2.4 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.6 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.1 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |