Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
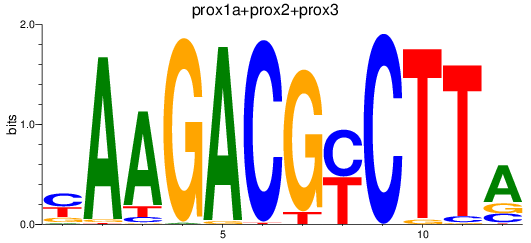
Results for prox1a+prox2+prox3
Z-value: 0.31
Transcription factors associated with prox1a+prox2+prox3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
prox2
|
ENSDARG00000041952 | prospero homeobox 2 |
|
prox1a
|
ENSDARG00000055158 | prospero homeobox 1a |
|
prox3
|
ENSDARG00000088810 | prospero homeobox 3 |
|
prox3
|
ENSDARG00000113014 | prospero homeobox 3 |
|
prox2
|
ENSDARG00000117137 | prospero homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| prox1a | dr11_v1_chr17_-_32865788_32865788 | -0.59 | 7.5e-03 | Click! |
| prox1b | dr11_v1_chr7_-_19940473_19940473 | -0.49 | 3.1e-02 | Click! |
| prox2 | dr11_v1_chr17_-_52521002_52521002 | -0.05 | 8.3e-01 | Click! |
Activity profile of prox1a+prox2+prox3 motif
Sorted Z-values of prox1a+prox2+prox3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_23219367 | 0.69 |
ENSDART00000003646
|
optc
|
opticin |
| chr13_+_7442023 | 0.64 |
ENSDART00000080975
|
tnfaip2b
|
tumor necrosis factor, alpha-induced protein 2b |
| chr4_+_5196469 | 0.58 |
ENSDART00000067386
|
rad51ap1
|
RAD51 associated protein 1 |
| chr14_+_31751260 | 0.52 |
ENSDART00000169796
|
si:dkeyp-11e3.1
|
si:dkeyp-11e3.1 |
| chr7_+_20917966 | 0.48 |
ENSDART00000129161
|
wrap53
|
WD repeat containing, antisense to TP53 |
| chr7_-_44604540 | 0.47 |
ENSDART00000149186
|
tk2
|
thymidine kinase 2, mitochondrial |
| chr22_+_10678141 | 0.47 |
ENSDART00000193341
|
hyal2b
|
hyaluronoglucosaminidase 2b |
| chr7_-_44605050 | 0.46 |
ENSDART00000148471
ENSDART00000149072 |
tk2
|
thymidine kinase 2, mitochondrial |
| chr17_-_106291 | 0.43 |
ENSDART00000160738
|
arhgap11a
|
Rho GTPase activating protein 11A |
| chr14_-_28473639 | 0.40 |
ENSDART00000173007
|
nxt2
|
nuclear transport factor 2-like export factor 2 |
| chr14_-_30971264 | 0.37 |
ENSDART00000010512
|
zgc:92907
|
zgc:92907 |
| chr5_-_61638125 | 0.37 |
ENSDART00000134314
|
si:dkey-261j4.3
|
si:dkey-261j4.3 |
| chr12_+_27117609 | 0.35 |
ENSDART00000076154
|
hoxb8b
|
homeobox B8b |
| chr25_+_16312258 | 0.35 |
ENSDART00000064187
|
parvaa
|
parvin, alpha a |
| chr13_+_47050726 | 0.34 |
ENSDART00000140045
|
anapc1
|
anaphase promoting complex subunit 1 |
| chr3_+_22335030 | 0.33 |
ENSDART00000055676
|
zgc:103564
|
zgc:103564 |
| chr16_+_42772678 | 0.32 |
ENSDART00000155575
|
si:ch211-135n15.2
|
si:ch211-135n15.2 |
| chr3_+_21200763 | 0.32 |
ENSDART00000067841
|
zgc:112038
|
zgc:112038 |
| chr23_+_21663631 | 0.31 |
ENSDART00000066125
|
dhrs3a
|
dehydrogenase/reductase (SDR family) member 3a |
| chr4_+_77973876 | 0.30 |
ENSDART00000057423
|
terfa
|
telomeric repeat binding factor a |
| chr7_-_38087865 | 0.29 |
ENSDART00000052366
|
cebpa
|
CCAAT/enhancer binding protein (C/EBP), alpha |
| chr12_+_6065661 | 0.29 |
ENSDART00000142418
|
sgms1
|
sphingomyelin synthase 1 |
| chr9_+_8968702 | 0.28 |
ENSDART00000008490
|
ube2al
|
ubiquitin conjugating enzyme E2 A, like |
| chr23_-_5759242 | 0.28 |
ENSDART00000055087
|
phlda3
|
pleckstrin homology-like domain, family A, member 3 |
| chr10_-_38316134 | 0.27 |
ENSDART00000149580
|
nrip1b
|
nuclear receptor interacting protein 1b |
| chr2_-_30055432 | 0.27 |
ENSDART00000056747
|
shhb
|
sonic hedgehog b |
| chr18_-_34549721 | 0.26 |
ENSDART00000137101
ENSDART00000021880 |
ssr3
|
signal sequence receptor, gamma |
| chr14_-_6286966 | 0.25 |
ENSDART00000168174
|
elp1
|
elongator complex protein 1 |
| chr5_+_27440325 | 0.25 |
ENSDART00000185815
ENSDART00000144013 |
loxl2b
|
lysyl oxidase-like 2b |
| chr16_+_51154918 | 0.25 |
ENSDART00000058290
|
dhdds
|
dehydrodolichyl diphosphate synthase |
| chr9_+_21401189 | 0.24 |
ENSDART00000062669
|
cx30.3
|
connexin 30.3 |
| chr15_-_23475051 | 0.24 |
ENSDART00000152460
|
nlrx1
|
NLR family member X1 |
| chr9_-_23033818 | 0.24 |
ENSDART00000022392
|
rnd3b
|
Rho family GTPase 3b |
| chr11_-_25461336 | 0.23 |
ENSDART00000014945
|
hcfc1a
|
host cell factor C1a |
| chr14_+_24840669 | 0.22 |
ENSDART00000106039
|
arhgef37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr13_+_11829072 | 0.21 |
ENSDART00000079356
ENSDART00000170160 |
sufu
|
suppressor of fused homolog (Drosophila) |
| chr13_-_35765028 | 0.21 |
ENSDART00000157391
|
erlec1
|
endoplasmic reticulum lectin 1 |
| chr10_-_8294965 | 0.20 |
ENSDART00000167380
|
plpp1a
|
phospholipid phosphatase 1a |
| chr1_-_59065217 | 0.20 |
ENSDART00000188972
|
CABZ01080042.1
|
|
| chr12_-_18898413 | 0.20 |
ENSDART00000181281
ENSDART00000121866 |
desi1b
|
desumoylating isopeptidase 1b |
| chr10_+_7719796 | 0.20 |
ENSDART00000191795
|
ggcx
|
gamma-glutamyl carboxylase |
| chr6_-_32411703 | 0.20 |
ENSDART00000151002
ENSDART00000078908 |
usp1
|
ubiquitin specific peptidase 1 |
| chr14_+_28473173 | 0.19 |
ENSDART00000017075
|
xiap
|
X-linked inhibitor of apoptosis |
| chr22_+_2198246 | 0.19 |
ENSDART00000165168
ENSDART00000157485 ENSDART00000168713 |
znf1157
znf1166
|
zinc finger protein 1157 zinc finger protein 1166 |
| chr5_+_13870340 | 0.19 |
ENSDART00000160690
|
hk2
|
hexokinase 2 |
| chr3_+_11072958 | 0.18 |
ENSDART00000158899
|
mrps7
|
mitochondrial ribosomal protein S7 |
| chr15_+_32711663 | 0.18 |
ENSDART00000157854
ENSDART00000167515 |
postnb
|
periostin, osteoblast specific factor b |
| chr19_+_19989380 | 0.18 |
ENSDART00000142841
|
osbpl3a
|
oxysterol binding protein-like 3a |
| chr3_-_15667713 | 0.18 |
ENSDART00000026658
|
zgc:66474
|
zgc:66474 |
| chr8_+_30600386 | 0.18 |
ENSDART00000164976
|
specc1la
|
sperm antigen with calponin homology and coiled-coil domains 1-like a |
| chr10_-_8295294 | 0.17 |
ENSDART00000075412
ENSDART00000163803 |
plpp1a
|
phospholipid phosphatase 1a |
| chr10_-_36253958 | 0.17 |
ENSDART00000126288
|
or110-2
|
odorant receptor, family D, subfamily 110, member 2 |
| chr10_+_29266392 | 0.16 |
ENSDART00000153577
|
crebzf
|
CREB/ATF bZIP transcription factor |
| chr21_-_303766 | 0.16 |
ENSDART00000150913
|
lhfpl2a
|
LHFPL tetraspan subfamily member 2a |
| chr19_+_42886413 | 0.16 |
ENSDART00000151298
|
arpp21
|
cAMP-regulated phosphoprotein, 21 |
| chr4_+_3455146 | 0.15 |
ENSDART00000180124
|
znf800b
|
zinc finger protein 800b |
| chr25_+_22587306 | 0.15 |
ENSDART00000067479
|
stra6
|
stimulated by retinoic acid 6 |
| chr23_+_36118738 | 0.15 |
ENSDART00000139319
|
hoxc5a
|
homeobox C5a |
| chr22_+_9239831 | 0.14 |
ENSDART00000133720
|
si:ch211-250k18.5
|
si:ch211-250k18.5 |
| chr22_+_9287929 | 0.14 |
ENSDART00000193522
|
si:ch211-250k18.7
|
si:ch211-250k18.7 |
| chr1_-_9486214 | 0.14 |
ENSDART00000137821
|
micall2b
|
mical-like 2b |
| chr2_+_2470687 | 0.14 |
ENSDART00000184024
ENSDART00000061955 |
myl13
|
myosin, light chain 13 |
| chr6_-_34006917 | 0.13 |
ENSDART00000190702
ENSDART00000184003 |
tie1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1 |
| chr22_-_23591493 | 0.13 |
ENSDART00000170266
|
f13b
|
coagulation factor XIII, B polypeptide |
| chr7_-_44604821 | 0.12 |
ENSDART00000148967
|
tk2
|
thymidine kinase 2, mitochondrial |
| chr20_+_192170 | 0.12 |
ENSDART00000189675
|
cx28.8
|
connexin 28.8 |
| chr5_-_51484156 | 0.12 |
ENSDART00000162064
|
CR388055.1
|
|
| chr8_+_11325310 | 0.12 |
ENSDART00000142577
|
fxn
|
frataxin |
| chr1_+_6161930 | 0.12 |
ENSDART00000037036
|
cnppd1
|
cyclin Pas1/PHO80 domain containing 1 |
| chr5_-_69707787 | 0.12 |
ENSDART00000108820
ENSDART00000149692 |
dguok
|
deoxyguanosine kinase |
| chr18_+_33264609 | 0.11 |
ENSDART00000050639
|
v2ra20
|
vomeronasal 2 receptor, a20 |
| chr23_+_16889352 | 0.10 |
ENSDART00000145275
|
si:dkey-147f3.8
|
si:dkey-147f3.8 |
| chr14_-_28566238 | 0.10 |
ENSDART00000172547
|
insb
|
preproinsulin b |
| chr9_-_42696408 | 0.10 |
ENSDART00000144744
|
col5a2a
|
collagen, type V, alpha 2a |
| chr19_+_7929704 | 0.10 |
ENSDART00000147015
|
si:dkey-266f7.4
|
si:dkey-266f7.4 |
| chr16_+_39196727 | 0.09 |
ENSDART00000017017
|
zdhhc3a
|
zinc finger, DHHC-type containing 3a |
| chr22_-_23591340 | 0.09 |
ENSDART00000167024
|
f13b
|
coagulation factor XIII, B polypeptide |
| chr4_+_3455665 | 0.09 |
ENSDART00000058277
|
znf800b
|
zinc finger protein 800b |
| chr13_+_35765317 | 0.08 |
ENSDART00000100156
ENSDART00000167650 |
agpat4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta) |
| chr25_-_10630496 | 0.08 |
ENSDART00000153639
ENSDART00000181722 ENSDART00000177834 |
ppp6r3
|
protein phosphatase 6, regulatory subunit 3 |
| chr19_-_13950231 | 0.08 |
ENSDART00000168665
|
sh3bgrl3
|
SH3 domain binding glutamate-rich protein like 3 |
| chr7_+_57866292 | 0.08 |
ENSDART00000138757
|
camk2d1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 1 |
| chr4_-_5913338 | 0.08 |
ENSDART00000183590
|
arl1
|
ADP-ribosylation factor-like 1 |
| chr16_-_13680692 | 0.07 |
ENSDART00000047452
|
ube2s
|
ubiquitin-conjugating enzyme E2S |
| chr5_+_37785152 | 0.07 |
ENSDART00000053511
ENSDART00000189812 |
myo1ca
|
myosin Ic, paralog a |
| chr15_+_37954666 | 0.06 |
ENSDART00000126534
ENSDART00000155548 |
si:dkey-238d18.7
|
si:dkey-238d18.7 |
| chr17_-_10059557 | 0.06 |
ENSDART00000092209
ENSDART00000161243 |
baz1a
|
bromodomain adjacent to zinc finger domain, 1A |
| chr10_+_6697369 | 0.06 |
ENSDART00000148606
|
si:ch211-57m13.8
|
si:ch211-57m13.8 |
| chr2_+_25658112 | 0.06 |
ENSDART00000051234
|
tnika
|
TRAF2 and NCK interacting kinase a |
| chr20_-_54014373 | 0.06 |
ENSDART00000152934
|
si:dkey-241l7.6
|
si:dkey-241l7.6 |
| chr20_-_40717900 | 0.06 |
ENSDART00000181663
|
cx43
|
connexin 43 |
| chr1_-_9485939 | 0.05 |
ENSDART00000157814
|
micall2b
|
mical-like 2b |
| chr16_+_11779761 | 0.05 |
ENSDART00000140297
|
si:dkey-250k15.4
|
si:dkey-250k15.4 |
| chr10_-_2713228 | 0.03 |
ENSDART00000123754
ENSDART00000126236 |
mier3a
|
mesoderm induction early response 1, family member 3 a |
| chr10_+_43037064 | 0.03 |
ENSDART00000160159
|
atg10
|
ATG10 autophagy related 10 homolog (S. cerevisiae) |
| chr17_-_31639845 | 0.02 |
ENSDART00000154196
|
znf839
|
zinc finger protein 839 |
| chr19_-_22621811 | 0.02 |
ENSDART00000090669
|
pleca
|
plectin a |
| chr19_-_21766461 | 0.02 |
ENSDART00000104279
|
znf516
|
zinc finger protein 516 |
| chr22_+_36692681 | 0.02 |
ENSDART00000186398
|
trim35-33
|
tripartite motif containing 35-33 |
| chr9_+_38684871 | 0.01 |
ENSDART00000147938
ENSDART00000004462 |
heg1
|
heart development protein with EGF-like domains 1 |
| chr7_+_7019911 | 0.01 |
ENSDART00000172421
|
rbm14b
|
RNA binding motif protein 14b |
| chr4_+_30788743 | 0.01 |
ENSDART00000184557
|
si:dkey-178j11.5
|
si:dkey-178j11.5 |
| chr4_-_75899294 | 0.01 |
ENSDART00000157887
|
si:dkey-261j11.3
|
si:dkey-261j11.3 |
| chr5_-_62306819 | 0.00 |
ENSDART00000168993
|
spata22
|
spermatogenesis associated 22 |
| chr13_-_16066997 | 0.00 |
ENSDART00000184790
|
SPATA48
|
spermatogenesis associated 48 |
| chr17_+_48164536 | 0.00 |
ENSDART00000161750
ENSDART00000156923 |
plekhd1
|
pleckstrin homology domain containing, family D (with coiled-coil domains) member 1 |
| chr23_-_13877898 | 0.00 |
ENSDART00000138696
|
g6pd
|
glucose-6-phosphate dehydrogenase |
| chr16_-_54919260 | 0.00 |
ENSDART00000156533
|
kdf1a
|
keratinocyte differentiation factor 1a |
| chr23_+_35672542 | 0.00 |
ENSDART00000046268
|
pmelb
|
premelanosome protein b |
| chr8_+_25616946 | 0.00 |
ENSDART00000133983
|
slc38a5a
|
solute carrier family 38, member 5a |
Network of associatons between targets according to the STRING database.
First level regulatory network of prox1a+prox2+prox3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0030575 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.1 | 0.3 | GO:0002926 | tRNA wobble base 5-methoxycarbonylmethyl-2-thiouridine biosynthesis.(GO:0002926) |
| 0.1 | 0.3 | GO:0032207 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) |
| 0.1 | 0.6 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.3 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.1 | 0.2 | GO:0046823 | regulation of protein import into nucleus(GO:0042306) negative regulation of protein import into nucleus(GO:0042308) negative regulation of nucleocytoplasmic transport(GO:0046823) negative regulation of protein localization to nucleus(GO:1900181) regulation of protein import(GO:1904589) negative regulation of protein import(GO:1904590) |
| 0.0 | 0.2 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.0 | 0.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.4 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.5 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.3 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.2 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.3 | GO:0060956 | cardiac endothelial cell differentiation(GO:0003348) endocardial cell differentiation(GO:0060956) |
| 0.0 | 0.2 | GO:0071938 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.0 | 0.3 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.3 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.1 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.2 | GO:0046546 | development of primary male sexual characteristics(GO:0046546) |
| 0.0 | 0.6 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.3 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.1 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.2 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.2 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.0 | 0.2 | GO:0016203 | muscle attachment(GO:0016203) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1904423 | dehydrodolichyl diphosphate synthase complex(GO:1904423) |
| 0.1 | 0.3 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.2 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.0 | 0.4 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.5 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.3 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.6 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.3 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.1 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 0.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 0.4 | GO:0000810 | diacylglycerol diphosphate phosphatase activity(GO:0000810) |
| 0.1 | 0.5 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.1 | 0.3 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 0.2 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.1 | 0.2 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.0 | 0.3 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.3 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.3 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.2 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.3 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.2 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.0 | 0.1 | GO:0019107 | myristoyltransferase activity(GO:0019107) |
| 0.0 | 0.1 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.0 | 0.2 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.3 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.2 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.3 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.5 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |