Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
Results for prdm14
Z-value: 0.46
Transcription factors associated with prdm14
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
prdm14
|
ENSDARG00000045371 | PR domain containing 14 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| prdm14 | dr11_v1_chr24_+_14451404_14451404 | -0.40 | 9.3e-02 | Click! |
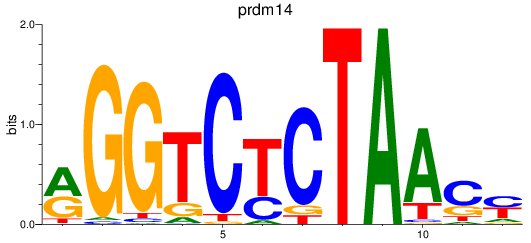
Activity profile of prdm14 motif
Sorted Z-values of prdm14 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_24135508 | 1.21 |
ENSDART00000171819
ENSDART00000103176 |
bcam
|
basal cell adhesion molecule (Lutheran blood group) |
| chr21_-_14630831 | 1.06 |
ENSDART00000132142
ENSDART00000089967 |
cacna1bb
|
calcium channel, voltage-dependent, N type, alpha 1B subunit, b |
| chr10_+_573667 | 1.04 |
ENSDART00000110384
|
smad4a
|
SMAD family member 4a |
| chr12_+_39685485 | 0.91 |
ENSDART00000163403
|
LO017650.1
|
|
| chr4_-_22207873 | 0.88 |
ENSDART00000142140
|
ppfia2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr21_+_44300689 | 0.88 |
ENSDART00000186298
ENSDART00000142810 |
gabra3
|
gamma-aminobutyric acid (GABA) A receptor, alpha 3 |
| chr23_+_39558508 | 0.82 |
ENSDART00000017902
|
camk1gb
|
calcium/calmodulin-dependent protein kinase IGb |
| chr12_-_6818676 | 0.76 |
ENSDART00000106391
|
pcdh15b
|
protocadherin-related 15b |
| chr21_-_21089781 | 0.73 |
ENSDART00000144361
|
ank1b
|
ankyrin 1, erythrocytic b |
| chr25_-_1235457 | 0.73 |
ENSDART00000093093
|
coro2bb
|
coronin, actin binding protein, 2Bb |
| chr3_-_18675688 | 0.73 |
ENSDART00000048218
|
slc5a11
|
solute carrier family 5 (sodium/inositol cotransporter), member 11 |
| chr5_-_69482891 | 0.71 |
ENSDART00000109487
|
CABZ01032476.1
|
|
| chr10_-_33343244 | 0.70 |
ENSDART00000164191
|
c2cd2
|
C2 calcium-dependent domain containing 2 |
| chr21_-_43949208 | 0.69 |
ENSDART00000150983
|
camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr19_-_4851411 | 0.62 |
ENSDART00000110398
|
fbxl20
|
F-box and leucine-rich repeat protein 20 |
| chr18_-_25646286 | 0.62 |
ENSDART00000099511
ENSDART00000186890 |
si:ch211-13k12.2
|
si:ch211-13k12.2 |
| chr2_-_32826108 | 0.58 |
ENSDART00000098834
|
prpf4ba
|
pre-mRNA processing factor 4Ba |
| chr9_-_22281854 | 0.56 |
ENSDART00000146319
|
crygm2d3
|
crystallin, gamma M2d3 |
| chr3_-_25268751 | 0.56 |
ENSDART00000139423
|
mgat3a
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase a |
| chr5_+_26079178 | 0.54 |
ENSDART00000145920
|
si:dkey-201c13.2
|
si:dkey-201c13.2 |
| chr18_+_44768829 | 0.52 |
ENSDART00000016271
|
ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr22_+_39074688 | 0.49 |
ENSDART00000153547
|
ip6k1
|
inositol hexakisphosphate kinase 1 |
| chr17_+_12075805 | 0.48 |
ENSDART00000155329
|
cnsta
|
consortin, connexin sorting protein a |
| chr8_+_35172594 | 0.48 |
ENSDART00000177146
|
BX897670.1
|
|
| chr20_-_51727860 | 0.47 |
ENSDART00000147044
|
brox
|
BRO1 domain and CAAX motif containing |
| chr17_+_25871304 | 0.47 |
ENSDART00000185143
|
wapla
|
WAPL cohesin release factor a |
| chr3_-_24456451 | 0.44 |
ENSDART00000024480
ENSDART00000156814 |
baiap2l2a
|
BAI1-associated protein 2-like 2a |
| chr5_-_67115872 | 0.41 |
ENSDART00000065262
|
rps6ka4
|
ribosomal protein S6 kinase, polypeptide 4 |
| chr15_-_42760110 | 0.39 |
ENSDART00000152490
|
si:ch211-181d7.3
|
si:ch211-181d7.3 |
| chr6_-_21988375 | 0.38 |
ENSDART00000161257
|
plxnb1b
|
plexin b1b |
| chr3_-_2593859 | 0.37 |
ENSDART00000143826
|
si:dkey-217f16.5
|
si:dkey-217f16.5 |
| chr8_-_49285801 | 0.35 |
ENSDART00000113159
|
prickle3
|
prickle homolog 3 |
| chr20_+_27020201 | 0.34 |
ENSDART00000126919
ENSDART00000016014 |
chga
|
chromogranin A |
| chr5_+_37903790 | 0.34 |
ENSDART00000162470
|
tmprss4b
|
transmembrane protease, serine 4b |
| chr12_-_36268723 | 0.34 |
ENSDART00000113740
|
kcnj16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr9_+_8364553 | 0.33 |
ENSDART00000190713
|
si:dkey-90l23.2
|
si:dkey-90l23.2 |
| chr13_+_49727333 | 0.32 |
ENSDART00000168799
ENSDART00000037559 |
ggps1
|
geranylgeranyl diphosphate synthase 1 |
| chr17_-_42492668 | 0.32 |
ENSDART00000183946
|
CR925755.2
|
|
| chr1_+_46509176 | 0.32 |
ENSDART00000166028
|
mcf2la
|
mcf.2 cell line derived transforming sequence-like a |
| chr13_-_31544365 | 0.31 |
ENSDART00000005670
|
dhrs7
|
dehydrogenase/reductase (SDR family) member 7 |
| chr14_-_30642819 | 0.27 |
ENSDART00000078154
|
npas4a
|
neuronal PAS domain protein 4a |
| chr17_+_27162367 | 0.26 |
ENSDART00000193345
|
rps6ka1
|
ribosomal protein S6 kinase a, polypeptide 1 |
| chr25_-_8030425 | 0.25 |
ENSDART00000014964
|
camk1db
|
calcium/calmodulin-dependent protein kinase 1Db |
| chr7_+_16806473 | 0.24 |
ENSDART00000113332
ENSDART00000173541 |
nav2a
|
neuron navigator 2a |
| chr25_-_8030113 | 0.23 |
ENSDART00000104674
|
camk1db
|
calcium/calmodulin-dependent protein kinase 1Db |
| chr22_+_2680895 | 0.23 |
ENSDART00000082245
|
zgc:194221
|
zgc:194221 |
| chr18_+_27926839 | 0.20 |
ENSDART00000191835
|
hipk3b
|
homeodomain interacting protein kinase 3b |
| chr7_+_13418812 | 0.20 |
ENSDART00000191905
ENSDART00000091567 |
dagla
|
diacylglycerol lipase, alpha |
| chr11_-_23080970 | 0.20 |
ENSDART00000127791
|
atp2b2
|
ATPase plasma membrane Ca2+ transporting 2 |
| chr7_-_17780048 | 0.19 |
ENSDART00000183336
|
si:dkey-106g10.7
|
si:dkey-106g10.7 |
| chr25_-_36248053 | 0.17 |
ENSDART00000134928
|
nfatc3b
|
nuclear factor of activated T cells 3b |
| chr16_-_26255877 | 0.17 |
ENSDART00000146214
|
erfl1
|
Ets2 repressor factor like 1 |
| chr15_+_19293744 | 0.17 |
ENSDART00000184994
ENSDART00000123815 |
jam3a
|
junctional adhesion molecule 3a |
| chr25_+_9003230 | 0.16 |
ENSDART00000180330
ENSDART00000142917 |
rag1
|
recombination activating gene 1 |
| chr6_+_518979 | 0.15 |
ENSDART00000151012
|
CACNA1I (1 of many)
|
si:ch73-379f7.5 |
| chr5_-_57898008 | 0.15 |
ENSDART00000050945
|
layna
|
layilin a |
| chr11_-_18791834 | 0.15 |
ENSDART00000156431
|
pfkfb2b
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2b |
| chr24_+_26402110 | 0.14 |
ENSDART00000133684
|
si:ch211-230g15.5
|
si:ch211-230g15.5 |
| chr12_-_4435303 | 0.13 |
ENSDART00000159007
|
si:ch211-173d10.1
|
si:ch211-173d10.1 |
| chr9_+_33261330 | 0.12 |
ENSDART00000135384
|
usp9
|
ubiquitin specific peptidase 9 |
| chr23_-_2448234 | 0.10 |
ENSDART00000082097
|
LO018513.1
|
|
| chr2_-_36819624 | 0.07 |
ENSDART00000140844
|
slitrk3b
|
SLIT and NTRK-like family, member 3b |
| chr5_-_45877387 | 0.06 |
ENSDART00000183714
ENSDART00000041503 |
slc4a4a
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4a |
| chr4_-_49069087 | 0.06 |
ENSDART00000150622
|
si:ch211-234c11.3
|
si:ch211-234c11.3 |
| chr7_+_51324834 | 0.06 |
ENSDART00000114429
|
usp12b
|
ubiquitin specific peptidase 12b |
| chr6_-_24384654 | 0.05 |
ENSDART00000164723
|
brdt
|
bromodomain, testis-specific |
| chr6_-_24143923 | 0.04 |
ENSDART00000157948
|
si:ch73-389b16.1
|
si:ch73-389b16.1 |
| chr15_-_21837207 | 0.03 |
ENSDART00000089953
|
sik2b
|
salt-inducible kinase 2b |
| chr8_-_25566347 | 0.03 |
ENSDART00000138289
ENSDART00000078022 |
prex1
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
| chr18_+_48793536 | 0.02 |
ENSDART00000168937
|
bmp16
|
bone morphogenetic protein 16 |
| chr13_+_4871886 | 0.01 |
ENSDART00000132301
|
micu1
|
mitochondrial calcium uptake 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of prdm14
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.2 | 0.5 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.1 | 0.5 | GO:0006549 | isoleucine metabolic process(GO:0006549) isoleucine biosynthetic process(GO:0009097) |
| 0.1 | 0.8 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 0.3 | GO:0090278 | negative regulation of peptide secretion(GO:0002792) negative regulation of peptide hormone secretion(GO:0090278) |
| 0.1 | 0.4 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.1 | 0.8 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 0.3 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.1 | 0.2 | GO:0098920 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.0 | 0.4 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.9 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.0 | 0.6 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 1.0 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 0.5 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.4 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.1 | GO:0001207 | histone displacement(GO:0001207) regulation of transcription involved in meiotic cell cycle(GO:0051037) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.0 | 0.2 | GO:0033151 | V(D)J recombination(GO:0033151) |
| 0.0 | 0.6 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 0.7 | GO:0003146 | heart jogging(GO:0003146) |
| 0.0 | 0.2 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0005948 | acetolactate synthase complex(GO:0005948) |
| 0.1 | 1.0 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.2 | GO:0097519 | DNA recombinase complex(GO:0097519) |
| 0.0 | 0.8 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.3 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.9 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.4 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 1.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0003984 | acetolactate synthase activity(GO:0003984) |
| 0.1 | 0.7 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 0.5 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.9 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 0.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 1.0 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 2.0 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.2 | GO:1990238 | double-stranded DNA endodeoxyribonuclease activity(GO:1990238) |
| 0.0 | 0.5 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.3 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 1.1 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.4 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.1 | 0.9 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.7 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.3 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.2 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |