Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
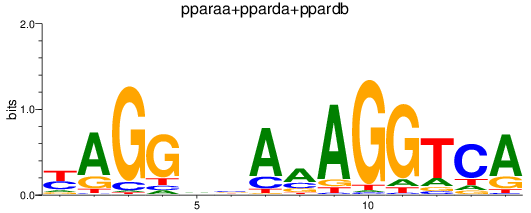
Results for pparaa+pparda+ppardb
Z-value: 0.48
Transcription factors associated with pparaa+pparda+ppardb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ppardb
|
ENSDARG00000009473 | peroxisome proliferator-activated receptor delta b |
|
pparaa
|
ENSDARG00000031777 | peroxisome proliferator-activated receptor alpha a |
|
pparda
|
ENSDARG00000044525 | peroxisome proliferator-activated receptor delta a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ppardb | dr11_v1_chr8_+_23703680_23703698 | 0.49 | 3.3e-02 | Click! |
| pparda | dr11_v1_chr22_+_997838_997838 | 0.35 | 1.5e-01 | Click! |
| pparaa | dr11_v1_chr4_-_18635005_18635005 | 0.21 | 3.9e-01 | Click! |
Activity profile of pparaa+pparda+ppardb motif
Sorted Z-values of pparaa+pparda+ppardb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_44027391 | 1.50 |
ENSDART00000145404
|
crybb1
|
crystallin, beta B1 |
| chr15_-_12500938 | 1.37 |
ENSDART00000159627
|
scn4ba
|
sodium channel, voltage-gated, type IV, beta a |
| chr15_+_45640906 | 1.23 |
ENSDART00000149361
ENSDART00000149079 |
sagb
|
S-antigen; retina and pineal gland (arrestin) b |
| chr6_+_60055168 | 1.08 |
ENSDART00000008752
|
pck1
|
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr22_+_38229321 | 1.05 |
ENSDART00000132670
ENSDART00000104504 |
si:ch211-284e20.8
|
si:ch211-284e20.8 |
| chr3_-_53486169 | 0.98 |
ENSDART00000115243
|
soul5
|
heme-binding protein soul5 |
| chr5_-_36549024 | 0.97 |
ENSDART00000097671
|
zgc:158432
|
zgc:158432 |
| chr23_-_39636195 | 0.95 |
ENSDART00000144439
|
vwa1
|
von Willebrand factor A domain containing 1 |
| chr5_-_23362602 | 0.88 |
ENSDART00000137120
|
gria3a
|
glutamate receptor, ionotropic, AMPA 3a |
| chr23_-_45504991 | 0.87 |
ENSDART00000148761
|
col24a1
|
collagen type XXIV alpha 1 |
| chr5_+_36974931 | 0.82 |
ENSDART00000193063
|
gjd1a
|
gap junction protein delta 1a |
| chr7_+_34297271 | 0.77 |
ENSDART00000180342
|
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr7_-_38633867 | 0.71 |
ENSDART00000137424
|
c1qtnf4
|
C1q and TNF related 4 |
| chr5_-_69940868 | 0.70 |
ENSDART00000185924
ENSDART00000097357 |
ugt2a4
|
UDP glucuronosyltransferase 2 family, polypeptide A4 |
| chr3_+_3810919 | 0.69 |
ENSDART00000056035
|
FQ311927.1
|
|
| chr3_+_30257582 | 0.68 |
ENSDART00000159497
ENSDART00000103457 ENSDART00000121883 |
mybpc2a
|
myosin binding protein C, fast type a |
| chr19_+_40861853 | 0.61 |
ENSDART00000126470
|
zgc:85777
|
zgc:85777 |
| chr10_-_17284055 | 0.56 |
ENSDART00000167464
|
GNAZ
|
G protein subunit alpha z |
| chr2_-_26476030 | 0.52 |
ENSDART00000145262
ENSDART00000132125 |
acadm
|
acyl-CoA dehydrogenase medium chain |
| chr3_+_32546499 | 0.51 |
ENSDART00000151739
|
pax10
|
paired box 10 |
| chr19_-_7043355 | 0.50 |
ENSDART00000104845
|
tapbp.1
|
TAP binding protein (tapasin), tandem duplicate 1 |
| chr15_+_47386939 | 0.49 |
ENSDART00000128224
|
FO904873.1
|
|
| chr7_-_24046999 | 0.48 |
ENSDART00000144616
ENSDART00000124653 ENSDART00000127813 |
dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr18_+_27515640 | 0.45 |
ENSDART00000181593
|
tp53i11b
|
tumor protein p53 inducible protein 11b |
| chr17_-_20711735 | 0.45 |
ENSDART00000150056
|
ank3b
|
ankyrin 3b |
| chr6_-_30863046 | 0.45 |
ENSDART00000155330
|
pde4ba
|
phosphodiesterase 4B, cAMP-specific a |
| chr21_+_15704556 | 0.44 |
ENSDART00000024858
ENSDART00000146909 |
chchd10
|
coiled-coil-helix-coiled-coil-helix domain containing 10 |
| chr19_+_7043634 | 0.43 |
ENSDART00000133954
|
mhc1uka
|
major histocompatibility complex class I UKA |
| chr12_+_21525496 | 0.42 |
ENSDART00000152974
|
ca10a
|
carbonic anhydrase Xa |
| chr11_+_30981777 | 0.40 |
ENSDART00000148949
|
cacna1ab
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit, b |
| chr21_+_26720803 | 0.39 |
ENSDART00000053797
|
slc3a2b
|
solute carrier family 3 (amino acid transporter heavy chain), member 2b |
| chr5_-_323712 | 0.39 |
ENSDART00000188793
|
HOOK3
|
hook microtubule tethering protein 3 |
| chr15_+_47618221 | 0.39 |
ENSDART00000168722
|
paf1
|
PAF1 homolog, Paf1/RNA polymerase II complex component |
| chr13_+_829585 | 0.37 |
ENSDART00000029051
|
gsta.2
|
glutathione S-transferase, alpha tandem duplicate 2 |
| chr2_-_32551178 | 0.37 |
ENSDART00000145603
|
smarcd3a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3a |
| chr17_+_12408188 | 0.36 |
ENSDART00000105218
|
khk
|
ketohexokinase |
| chr20_-_48604199 | 0.35 |
ENSDART00000161762
ENSDART00000170894 |
mgst3a
|
microsomal glutathione S-transferase 3a |
| chr25_-_35113891 | 0.34 |
ENSDART00000190724
|
zgc:165555
|
zgc:165555 |
| chr21_-_19006631 | 0.34 |
ENSDART00000080269
ENSDART00000191682 |
pgam2
|
phosphoglycerate mutase 2 (muscle) |
| chr25_-_19420949 | 0.34 |
ENSDART00000181338
|
map1ab
|
microtubule-associated protein 1Ab |
| chr17_+_30450163 | 0.34 |
ENSDART00000104257
|
lpin1
|
lipin 1 |
| chr11_+_11201096 | 0.34 |
ENSDART00000171916
ENSDART00000171521 ENSDART00000087105 ENSDART00000159603 |
myom2a
|
myomesin 2a |
| chr5_+_38276582 | 0.33 |
ENSDART00000158532
|
gnb2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr7_+_34296789 | 0.33 |
ENSDART00000052471
ENSDART00000173798 ENSDART00000173778 |
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr1_-_46859398 | 0.32 |
ENSDART00000135795
|
agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr9_+_32069989 | 0.32 |
ENSDART00000139540
|
si:dkey-83m22.7
|
si:dkey-83m22.7 |
| chr12_-_31457301 | 0.32 |
ENSDART00000043887
ENSDART00000148603 |
acsl5
|
acyl-CoA synthetase long chain family member 5 |
| chr7_+_11459235 | 0.31 |
ENSDART00000159611
|
il16
|
interleukin 16 |
| chr9_+_41224100 | 0.30 |
ENSDART00000141792
|
stat1b
|
signal transducer and activator of transcription 1b |
| chr5_-_30984010 | 0.29 |
ENSDART00000182367
|
spns3
|
spinster homolog 3 (Drosophila) |
| chr9_+_54686686 | 0.28 |
ENSDART00000066198
|
rab9a
|
RAB9A, member RAS oncogene family |
| chr13_+_28819768 | 0.27 |
ENSDART00000191401
ENSDART00000188895 ENSDART00000101653 |
CU639469.1
|
|
| chr17_+_12408405 | 0.27 |
ENSDART00000154827
ENSDART00000048440 ENSDART00000156429 |
khk
|
ketohexokinase |
| chr6_-_59563597 | 0.26 |
ENSDART00000166311
|
INHBE
|
inhibin beta E subunit |
| chr12_+_7491690 | 0.26 |
ENSDART00000152564
|
phyhiplb
|
phytanoyl-CoA 2-hydroxylase interacting protein-like b |
| chr12_+_46708920 | 0.26 |
ENSDART00000153089
|
exoc7
|
exocyst complex component 7 |
| chr1_-_17797802 | 0.26 |
ENSDART00000041215
|
sorbs2a
|
sorbin and SH3 domain containing 2a |
| chr18_+_31410652 | 0.25 |
ENSDART00000098504
|
def8
|
differentially expressed in FDCP 8 homolog (mouse) |
| chr20_-_7128612 | 0.24 |
ENSDART00000146755
ENSDART00000036871 |
si:ch211-121a2.4
|
si:ch211-121a2.4 |
| chr12_-_6033824 | 0.24 |
ENSDART00000131301
ENSDART00000139419 ENSDART00000032050 |
g6pca.1
|
glucose-6-phosphatase a, catalytic subunit, tandem duplicate 1 |
| chr24_+_2961098 | 0.24 |
ENSDART00000163760
ENSDART00000170835 |
eci2
|
enoyl-CoA delta isomerase 2 |
| chr22_+_18469004 | 0.24 |
ENSDART00000061430
|
cilp2
|
cartilage intermediate layer protein 2 |
| chr14_+_1719367 | 0.24 |
ENSDART00000157696
|
trpc7b
|
transient receptor potential cation channel, subfamily C, member 7b |
| chr1_+_40802454 | 0.23 |
ENSDART00000193568
|
cpz
|
carboxypeptidase Z |
| chr6_-_39764995 | 0.23 |
ENSDART00000085277
|
pfkmb
|
phosphofructokinase, muscle b |
| chr19_-_43552252 | 0.23 |
ENSDART00000138308
|
gpr186
|
G protein-coupled receptor 186 |
| chr8_-_2529878 | 0.22 |
ENSDART00000056767
|
acaa2
|
acetyl-CoA acyltransferase 2 |
| chr5_-_30984271 | 0.21 |
ENSDART00000051392
|
spns3
|
spinster homolog 3 (Drosophila) |
| chr15_-_6946286 | 0.20 |
ENSDART00000019330
|
ech1
|
enoyl CoA hydratase 1, peroxisomal |
| chr22_-_10774735 | 0.20 |
ENSDART00000081156
|
timm13
|
translocase of inner mitochondrial membrane 13 homolog (yeast) |
| chr4_+_60492313 | 0.20 |
ENSDART00000191043
ENSDART00000191631 |
si:dkey-211i20.2
|
si:dkey-211i20.2 |
| chr20_-_26937453 | 0.20 |
ENSDART00000139756
|
ftr97
|
finTRIM family, member 97 |
| chr1_-_35113974 | 0.19 |
ENSDART00000192811
ENSDART00000167461 |
BX897691.1
|
|
| chr25_+_36347126 | 0.19 |
ENSDART00000152449
|
si:ch211-113a14.22
|
si:ch211-113a14.22 |
| chr20_-_48604621 | 0.18 |
ENSDART00000161769
ENSDART00000157871 |
mgst3a
|
microsomal glutathione S-transferase 3a |
| chr13_+_43050562 | 0.18 |
ENSDART00000016602
|
cdh23
|
cadherin-related 23 |
| chr15_+_28268135 | 0.18 |
ENSDART00000152536
ENSDART00000188550 |
myo1cb
|
myosin Ic, paralog b |
| chr24_-_29586082 | 0.16 |
ENSDART00000136763
|
vav3a
|
vav 3 guanine nucleotide exchange factor a |
| chr20_-_47953524 | 0.15 |
ENSDART00000167986
|
HADHB
|
hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit beta |
| chr6_-_39765546 | 0.15 |
ENSDART00000185767
|
pfkmb
|
phosphofructokinase, muscle b |
| chr12_-_5120339 | 0.15 |
ENSDART00000168759
|
rbp4
|
retinol binding protein 4, plasma |
| chr12_-_44199316 | 0.15 |
ENSDART00000170378
|
si:ch73-329n5.1
|
si:ch73-329n5.1 |
| chr20_+_1088658 | 0.14 |
ENSDART00000162991
|
BX537249.1
|
|
| chr15_+_31820536 | 0.14 |
ENSDART00000045921
|
frya
|
furry homolog a (Drosophila) |
| chr12_-_5120175 | 0.14 |
ENSDART00000160729
|
rbp4
|
retinol binding protein 4, plasma |
| chr20_+_43379029 | 0.14 |
ENSDART00000142486
ENSDART00000186486 |
unc93a
|
unc-93 homolog A |
| chr22_+_9069081 | 0.14 |
ENSDART00000187842
|
BX248395.1
|
|
| chr3_-_18805225 | 0.13 |
ENSDART00000133471
ENSDART00000131758 |
msrb1a
|
methionine sulfoxide reductase B1a |
| chr20_+_54738210 | 0.13 |
ENSDART00000151399
|
pak7
|
p21 protein (Cdc42/Rac)-activated kinase 7 |
| chr19_+_19652439 | 0.13 |
ENSDART00000165934
|
hibadha
|
3-hydroxyisobutyrate dehydrogenase a |
| chr20_+_47953047 | 0.12 |
ENSDART00000079734
|
hadhaa
|
hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha a |
| chr4_-_20243021 | 0.11 |
ENSDART00000048023
|
cacna2d4a
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4a |
| chr18_+_13315739 | 0.11 |
ENSDART00000143404
|
si:ch211-260p9.3
|
si:ch211-260p9.3 |
| chr5_-_66301142 | 0.11 |
ENSDART00000067541
|
prlrb
|
prolactin receptor b |
| chr7_+_39011852 | 0.11 |
ENSDART00000093009
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr5_+_58421536 | 0.10 |
ENSDART00000173410
|
ccdc15
|
coiled-coil domain containing 15 |
| chr17_-_12196865 | 0.10 |
ENSDART00000154694
|
kif28
|
kinesin family member 28 |
| chr8_-_979735 | 0.10 |
ENSDART00000149612
|
znf366
|
zinc finger protein 366 |
| chr6_-_28117995 | 0.10 |
ENSDART00000147253
|
si:ch73-194h10.3
|
si:ch73-194h10.3 |
| chr6_+_154556 | 0.10 |
ENSDART00000193153
|
zglp1
|
zinc finger, GATA-like protein 1 |
| chr10_+_45128375 | 0.10 |
ENSDART00000164805
|
camk2b2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 2 |
| chr6_+_43353271 | 0.10 |
ENSDART00000191775
|
BX927362.2
|
|
| chr1_+_58260886 | 0.09 |
ENSDART00000110453
|
si:dkey-222h21.8
|
si:dkey-222h21.8 |
| chr1_+_58442694 | 0.09 |
ENSDART00000160897
|
zgc:194906
|
zgc:194906 |
| chr22_-_38274188 | 0.09 |
ENSDART00000139420
ENSDART00000015117 |
elavl2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr17_+_30545895 | 0.09 |
ENSDART00000076739
|
nhsl1a
|
NHS-like 1a |
| chr17_+_450956 | 0.09 |
ENSDART00000183022
ENSDART00000171386 |
zgc:194887
|
zgc:194887 |
| chr7_-_29231686 | 0.09 |
ENSDART00000173780
|
herc1
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
| chr4_+_67999732 | 0.08 |
ENSDART00000190597
|
si:ch211-133h13.1
|
si:ch211-133h13.1 |
| chr20_-_24443680 | 0.08 |
ENSDART00000191337
|
CR385053.3
|
|
| chr1_+_58139102 | 0.08 |
ENSDART00000134826
|
si:ch211-15j1.4
|
si:ch211-15j1.4 |
| chr13_-_31878263 | 0.07 |
ENSDART00000180411
|
syt14a
|
synaptotagmin XIVa |
| chr13_-_31878043 | 0.07 |
ENSDART00000187720
|
syt14a
|
synaptotagmin XIVa |
| chr1_+_58353661 | 0.07 |
ENSDART00000140074
|
si:dkey-222h21.2
|
si:dkey-222h21.2 |
| chr6_+_60036767 | 0.07 |
ENSDART00000155009
|
tgm8
|
transglutaminase 8 |
| chr5_+_38200807 | 0.07 |
ENSDART00000100769
|
hsd20b2
|
hydroxysteroid (20-beta) dehydrogenase 2 |
| chr19_+_32158010 | 0.07 |
ENSDART00000005255
|
mrpl53
|
mitochondrial ribosomal protein L53 |
| chr20_+_15015557 | 0.07 |
ENSDART00000039345
|
myoc
|
myocilin |
| chr18_+_44769027 | 0.07 |
ENSDART00000145190
|
ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr5_-_50600512 | 0.06 |
ENSDART00000190318
|
mctp1a
|
multiple C2 domains, transmembrane 1a |
| chr1_+_57050899 | 0.06 |
ENSDART00000152601
|
si:ch211-1f22.14
|
si:ch211-1f22.14 |
| chr10_+_15340768 | 0.06 |
ENSDART00000046274
ENSDART00000168909 |
trappc13
|
trafficking protein particle complex 13 |
| chr4_-_50926767 | 0.06 |
ENSDART00000183430
|
si:ch211-208f21.3
|
si:ch211-208f21.3 |
| chr12_-_684200 | 0.06 |
ENSDART00000152122
|
si:ch211-176g6.2
|
si:ch211-176g6.2 |
| chr18_+_15182965 | 0.05 |
ENSDART00000137018
|
si:ch73-62b13.1
|
si:ch73-62b13.1 |
| chr4_+_75577480 | 0.05 |
ENSDART00000188196
|
si:ch211-227e10.1
|
si:ch211-227e10.1 |
| chr13_+_28821841 | 0.05 |
ENSDART00000179900
|
CU639469.1
|
|
| chr11_+_24957858 | 0.05 |
ENSDART00000145647
|
sulf2a
|
sulfatase 2a |
| chr20_-_38836161 | 0.04 |
ENSDART00000061358
|
si:dkey-221h15.4
|
si:dkey-221h15.4 |
| chr6_-_41138854 | 0.03 |
ENSDART00000128723
ENSDART00000151055 ENSDART00000132484 |
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr4_-_36032177 | 0.02 |
ENSDART00000170143
|
zgc:174180
|
zgc:174180 |
| chr18_-_3456117 | 0.02 |
ENSDART00000158508
|
myo7aa
|
myosin VIIAa |
| chr16_-_29480335 | 0.02 |
ENSDART00000148930
|
lingo4b
|
leucine rich repeat and Ig domain containing 4b |
| chr14_+_24258562 | 0.02 |
ENSDART00000106074
ENSDART00000173227 |
chrm1a
|
cholinergic receptor, muscarinic 1a |
| chr23_+_36340520 | 0.02 |
ENSDART00000011201
|
copz1
|
coatomer protein complex, subunit zeta 1 |
| chr22_+_9287929 | 0.01 |
ENSDART00000193522
|
si:ch211-250k18.7
|
si:ch211-250k18.7 |
| chr24_-_7587401 | 0.01 |
ENSDART00000093163
|
galnt11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr18_+_16744307 | 0.01 |
ENSDART00000179872
ENSDART00000133490 |
lyve1b
|
lymphatic vessel endothelial hyaluronic receptor 1b |
| chr18_+_27926839 | 0.01 |
ENSDART00000191835
|
hipk3b
|
homeodomain interacting protein kinase 3b |
| chr22_+_9114416 | 0.01 |
ENSDART00000190169
|
nlrp15
|
NACHT, LRR and PYD domains-containing protein 15 |
| chr7_+_48675347 | 0.01 |
ENSDART00000157917
ENSDART00000185580 |
trpm5
|
transient receptor potential cation channel, subfamily M, member 5 |
| chr20_-_26936887 | 0.00 |
ENSDART00000160827
|
ftr79
|
finTRIM family, member 79 |
| chr16_+_28601974 | 0.00 |
ENSDART00000018235
|
crot
|
carnitine O-octanoyltransferase |
Network of associatons between targets according to the STRING database.
First level regulatory network of pparaa+pparda+ppardb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0019626 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) short-chain fatty acid catabolic process(GO:0019626) response to dexamethasone(GO:0071548) |
| 0.2 | 0.6 | GO:0009750 | response to fructose(GO:0009750) |
| 0.2 | 1.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 0.9 | GO:0038065 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.1 | 0.3 | GO:0010747 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.1 | 1.2 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.1 | 0.5 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) medium-chain fatty acid catabolic process(GO:0051793) |
| 0.0 | 0.5 | GO:0019883 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.4 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.3 | GO:0034633 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.0 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.2 | GO:0035678 | neuromast hair cell morphogenesis(GO:0035678) |
| 0.0 | 0.4 | GO:0060142 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) |
| 0.0 | 0.4 | GO:0006735 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.3 | GO:0060337 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.5 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.2 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 0.1 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.3 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.4 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.4 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.1 | GO:0009097 | isoleucine metabolic process(GO:0006549) isoleucine biosynthetic process(GO:0009097) |
| 0.0 | 0.3 | GO:0050926 | regulation of positive chemotaxis(GO:0050926) positive regulation of positive chemotaxis(GO:0050927) induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.3 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.4 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.0 | 0.3 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.1 | 0.4 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 0.2 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 0.8 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 1.2 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.3 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.4 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0005948 | acetolactate synthase complex(GO:0005948) |
| 0.0 | 0.9 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.1 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.0 | 0.2 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.3 | GO:0031430 | M band(GO:0031430) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 0.3 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 1.1 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 0.2 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.1 | 0.3 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.1 | 0.8 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 0.9 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.3 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.4 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.2 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.1 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.0 | 0.1 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.0 | 0.3 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 1.1 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.9 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 1.1 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.3 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.1 | GO:0003984 | acetolactate synthase activity(GO:0003984) |
| 0.0 | 0.6 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 1.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.4 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.2 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 1.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.9 | NABA COLLAGENS | Genes encoding collagen proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.9 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |