Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
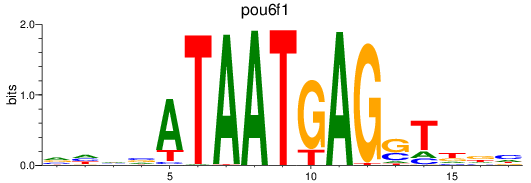
Results for pou6f1
Z-value: 0.53
Transcription factors associated with pou6f1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pou6f1
|
ENSDARG00000011570 | POU class 6 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pou6f1 | dr11_v1_chr23_-_33702900_33702900 | -0.60 | 7.0e-03 | Click! |
Activity profile of pou6f1 motif
Sorted Z-values of pou6f1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_22339582 | 3.26 |
ENSDART00000134805
|
crygm2d1
|
crystallin, gamma M2d1 |
| chr16_+_26017360 | 2.05 |
ENSDART00000149466
|
prss59.2
|
protease, serine, 59, tandem duplicate 2 |
| chr16_+_26012569 | 1.90 |
ENSDART00000148846
|
prss59.1
|
protease, serine, 59, tandem duplicate 1 |
| chr14_+_50770537 | 1.31 |
ENSDART00000158723
|
sncb
|
synuclein, beta |
| chr15_-_16098531 | 1.18 |
ENSDART00000080377
|
aldoca
|
aldolase C, fructose-bisphosphate, a |
| chr4_+_22365061 | 1.00 |
ENSDART00000039277
|
lhfpl3
|
LHFPL tetraspan subfamily member 3 |
| chr3_-_28250722 | 0.99 |
ENSDART00000165936
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr6_+_60055168 | 0.85 |
ENSDART00000008752
|
pck1
|
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr12_+_20352400 | 0.80 |
ENSDART00000066383
|
hbae5
|
hemoglobin, alpha embryonic 5 |
| chr22_-_15385442 | 0.79 |
ENSDART00000090975
|
tmem264
|
transmembrane protein 264 |
| chr16_-_11798994 | 0.77 |
ENSDART00000135408
|
cnfn
|
cornifelin |
| chr11_+_39672874 | 0.76 |
ENSDART00000046663
ENSDART00000157659 |
camta1b
|
calmodulin binding transcription activator 1b |
| chr3_+_33341640 | 0.74 |
ENSDART00000186352
|
pyya
|
peptide YYa |
| chr24_-_32408404 | 0.74 |
ENSDART00000144157
|
si:ch211-56a11.2
|
si:ch211-56a11.2 |
| chr4_-_72609735 | 0.72 |
ENSDART00000174299
ENSDART00000159227 |
si:cabz01054394.6
|
si:cabz01054394.6 |
| chr2_+_9552456 | 0.71 |
ENSDART00000056896
|
dnajb4
|
DnaJ (Hsp40) homolog, subfamily B, member 4 |
| chr1_+_50968908 | 0.67 |
ENSDART00000150353
ENSDART00000012842 |
mdh1aa
|
malate dehydrogenase 1Aa, NAD (soluble) |
| chr21_+_25231160 | 0.65 |
ENSDART00000063089
ENSDART00000139127 |
gng8
|
guanine nucleotide binding protein (G protein), gamma 8 |
| chr19_-_5103313 | 0.63 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr20_+_15982482 | 0.61 |
ENSDART00000020999
|
angptl1a
|
angiopoietin-like 1a |
| chr4_+_14826299 | 0.57 |
ENSDART00000067035
|
shisal1a
|
shisa like 1a |
| chr16_-_5154024 | 0.57 |
ENSDART00000132069
ENSDART00000060635 |
dctn3
|
dynactin 3 (p22) |
| chr8_+_37168570 | 0.54 |
ENSDART00000098636
|
csf1b
|
colony stimulating factor 1b (macrophage) |
| chr4_+_12612723 | 0.51 |
ENSDART00000133767
|
lmo3
|
LIM domain only 3 |
| chr18_-_1185772 | 0.50 |
ENSDART00000143245
|
nptnb
|
neuroplastin b |
| chr1_+_45121393 | 0.50 |
ENSDART00000142702
|
muc13a
|
mucin 13a, cell surface associated |
| chr17_+_12698532 | 0.48 |
ENSDART00000064509
ENSDART00000136830 |
stmn4l
|
stathmin-like 4, like |
| chr16_-_2414063 | 0.48 |
ENSDART00000073621
|
zgc:152945
|
zgc:152945 |
| chr12_-_28983584 | 0.48 |
ENSDART00000112374
|
zgc:171713
|
zgc:171713 |
| chr7_+_38717624 | 0.47 |
ENSDART00000132522
|
syt13
|
synaptotagmin XIII |
| chr9_-_44953349 | 0.47 |
ENSDART00000135156
|
vil1
|
villin 1 |
| chr9_-_44953664 | 0.46 |
ENSDART00000188558
ENSDART00000185210 |
vil1
|
villin 1 |
| chr19_+_19600297 | 0.45 |
ENSDART00000160134
ENSDART00000183493 |
hibadha
|
3-hydroxyisobutyrate dehydrogenase a |
| chr12_+_18681477 | 0.43 |
ENSDART00000127981
ENSDART00000143979 |
rgs9b
|
regulator of G protein signaling 9b |
| chr24_-_21404367 | 0.43 |
ENSDART00000152093
|
atp8a2
|
ATPase phospholipid transporting 8A2 |
| chr22_+_34710473 | 0.43 |
ENSDART00000155906
|
hoga1
|
4-hydroxy-2-oxoglutarate aldolase 1 |
| chr21_-_26918901 | 0.42 |
ENSDART00000100685
|
lrfn4a
|
leucine rich repeat and fibronectin type III domain containing 4a |
| chr9_+_11034314 | 0.41 |
ENSDART00000032695
|
asic4a
|
acid-sensing (proton-gated) ion channel family member 4a |
| chr5_+_64732270 | 0.41 |
ENSDART00000134241
|
olfm1a
|
olfactomedin 1a |
| chr19_+_42469058 | 0.41 |
ENSDART00000076915
|
si:dkey-166k12.1
|
si:dkey-166k12.1 |
| chr4_+_12615836 | 0.37 |
ENSDART00000003583
|
lmo3
|
LIM domain only 3 |
| chr9_-_32672129 | 0.34 |
ENSDART00000140581
|
gzm3.4
|
granzyme 3, tandem duplicate 4 |
| chr9_-_6624663 | 0.34 |
ENSDART00000092537
|
GPR45
|
si:dkeyp-118h3.5 |
| chr1_+_25801648 | 0.33 |
ENSDART00000129471
|
gucy1b1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr3_+_52753416 | 0.31 |
ENSDART00000171185
|
gmip
|
GEM interacting protein |
| chr25_+_6186823 | 0.30 |
ENSDART00000153526
|
oaz2a
|
ornithine decarboxylase antizyme 2a |
| chr25_+_11002640 | 0.29 |
ENSDART00000156646
ENSDART00000073383 |
mhc1lga
|
major histocompatibility complex class I LGA |
| chr2_-_21349425 | 0.28 |
ENSDART00000171699
|
hhatla
|
hedgehog acyltransferase like, a |
| chr19_+_30662529 | 0.25 |
ENSDART00000175662
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr25_-_28600433 | 0.25 |
ENSDART00000138980
|
agbl2
|
ATP/GTP binding protein-like 2 |
| chr4_+_359970 | 0.25 |
ENSDART00000139832
|
tmem181
|
transmembrane protein 181 |
| chr24_+_41931585 | 0.24 |
ENSDART00000130310
|
epb41l3a
|
erythrocyte membrane protein band 4.1-like 3a |
| chr7_-_19364813 | 0.24 |
ENSDART00000173977
|
ntn4
|
netrin 4 |
| chr19_-_8600383 | 0.23 |
ENSDART00000081546
|
trim46b
|
tripartite motif containing 46b |
| chr13_+_25434932 | 0.23 |
ENSDART00000128562
|
si:dkey-51a16.9
|
si:dkey-51a16.9 |
| chr4_-_18202291 | 0.23 |
ENSDART00000169704
|
anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr9_+_41612642 | 0.22 |
ENSDART00000138473
|
spegb
|
SPEG complex locus b |
| chr19_+_46222918 | 0.22 |
ENSDART00000158703
|
vps28
|
vacuolar protein sorting 28 (yeast) |
| chr11_-_7320211 | 0.22 |
ENSDART00000091664
|
apc2
|
adenomatosis polyposis coli 2 |
| chr1_+_19649545 | 0.21 |
ENSDART00000054575
|
tmem192
|
transmembrane protein 192 |
| chr2_-_21625493 | 0.21 |
ENSDART00000137169
|
zgc:55781
|
zgc:55781 |
| chr19_+_46222428 | 0.20 |
ENSDART00000183984
|
vps28
|
vacuolar protein sorting 28 (yeast) |
| chr13_-_21701323 | 0.20 |
ENSDART00000164112
|
si:dkey-191g9.7
|
si:dkey-191g9.7 |
| chr16_-_24044664 | 0.19 |
ENSDART00000136982
|
bcam
|
basal cell adhesion molecule (Lutheran blood group) |
| chr2_-_57991041 | 0.18 |
ENSDART00000111512
|
si:dkeyp-68b7.12
|
si:dkeyp-68b7.12 |
| chr2_+_29249204 | 0.18 |
ENSDART00000168957
|
cdh18a
|
cadherin 18, type 2a |
| chr2_-_57941037 | 0.18 |
ENSDART00000131420
|
si:dkeyp-68b7.5
|
si:dkeyp-68b7.5 |
| chr22_+_34710036 | 0.16 |
ENSDART00000025820
|
hoga1
|
4-hydroxy-2-oxoglutarate aldolase 1 |
| chr20_-_10288156 | 0.16 |
ENSDART00000064110
|
si:dkey-63b1.1
|
si:dkey-63b1.1 |
| chr17_-_2817035 | 0.14 |
ENSDART00000152002
|
gpr65
|
G protein-coupled receptor 65 |
| chr2_+_29249561 | 0.14 |
ENSDART00000099157
|
cdh18a
|
cadherin 18, type 2a |
| chr22_+_1462997 | 0.14 |
ENSDART00000157476
|
si:dkeyp-53d3.5
|
si:dkeyp-53d3.5 |
| chr13_-_42560662 | 0.14 |
ENSDART00000124898
|
CR792417.1
|
|
| chr5_-_35953472 | 0.14 |
ENSDART00000143448
|
rxfp2l
|
relaxin/insulin-like family peptide receptor 2, like |
| chr2_-_20981907 | 0.13 |
ENSDART00000113384
|
lyrm4
|
LYR motif containing 4 |
| chr25_+_37480285 | 0.13 |
ENSDART00000166187
|
CABZ01095001.1
|
|
| chr15_-_33172246 | 0.13 |
ENSDART00000158666
|
nbeab
|
neurobeachin b |
| chr4_-_76370630 | 0.13 |
ENSDART00000168831
ENSDART00000174313 |
si:ch73-158p21.3
|
si:ch73-158p21.3 |
| chr2_-_50372467 | 0.13 |
ENSDART00000108900
|
cntnap2b
|
contactin associated protein like 2b |
| chr19_-_9503473 | 0.13 |
ENSDART00000091615
|
iffo1a
|
intermediate filament family orphan 1a |
| chr14_+_46003973 | 0.12 |
ENSDART00000145241
ENSDART00000141545 |
slu7
|
SLU7 homolog, splicing factor |
| chr21_+_40292801 | 0.12 |
ENSDART00000174220
|
si:ch211-218m3.16
|
si:ch211-218m3.16 |
| chr3_+_17689419 | 0.12 |
ENSDART00000192842
ENSDART00000126029 |
dhx58
|
DEXH (Asp-Glu-X-His) box polypeptide 58 |
| chr12_+_32729470 | 0.12 |
ENSDART00000175712
|
rbfox3a
|
RNA binding fox-1 homolog 3a |
| chr1_+_40308077 | 0.12 |
ENSDART00000138992
|
vwa10.2
|
von Willebrand factor A domain containing 10, tandem duplicate 2 |
| chr16_-_21038015 | 0.12 |
ENSDART00000059239
|
snx10b
|
sorting nexin 10b |
| chr22_-_11520405 | 0.11 |
ENSDART00000063157
|
slc26a11
|
solute carrier family 26 (anion exchanger), member 11 |
| chr4_-_72609118 | 0.11 |
ENSDART00000174077
|
si:cabz01054396.2
|
si:cabz01054396.2 |
| chr16_-_25368048 | 0.10 |
ENSDART00000132445
|
rbfa
|
ribosome binding factor A |
| chr18_+_50523870 | 0.10 |
ENSDART00000151593
|
ubl7b
|
ubiquitin-like 7b (bone marrow stromal cell-derived) |
| chr11_+_33628104 | 0.10 |
ENSDART00000165318
|
thsd7bb
|
thrombospondin, type I, domain containing 7Bb |
| chr4_-_19028861 | 0.10 |
ENSDART00000166374
|
si:dkey-31f5.11
|
si:dkey-31f5.11 |
| chr21_-_20733615 | 0.09 |
ENSDART00000145544
|
si:ch211-22d5.2
|
si:ch211-22d5.2 |
| chr4_+_47751483 | 0.09 |
ENSDART00000160043
|
si:ch211-196f19.1
|
si:ch211-196f19.1 |
| chr11_+_29770966 | 0.09 |
ENSDART00000088624
ENSDART00000124471 |
rpgrb
|
retinitis pigmentosa GTPase regulator b |
| chr13_-_46200240 | 0.09 |
ENSDART00000056984
|
ftr69
|
finTRIM family, member 69 |
| chr2_+_38892631 | 0.08 |
ENSDART00000134040
|
si:ch211-119o8.4
|
si:ch211-119o8.4 |
| chr20_-_51697437 | 0.08 |
ENSDART00000145391
|
si:ch211-14a11.2
|
si:ch211-14a11.2 |
| chr19_-_5265155 | 0.07 |
ENSDART00000145003
|
prf1.3
|
perforin 1.3 |
| chr25_-_1333676 | 0.06 |
ENSDART00000183182
ENSDART00000188907 |
cln6b
|
CLN6, transmembrane ER protein b |
| chr23_+_45860676 | 0.06 |
ENSDART00000012234
|
syap1
|
synapse associated protein 1 |
| chr19_-_23799729 | 0.06 |
ENSDART00000151913
|
pimr88
|
Pim proto-oncogene, serine/threonine kinase, related 88 |
| chr24_-_16979728 | 0.06 |
ENSDART00000005331
|
klhl15
|
kelch-like family member 15 |
| chr1_+_44767657 | 0.06 |
ENSDART00000073717
|
si:dkey-9i23.4
|
si:dkey-9i23.4 |
| chr6_-_29105727 | 0.06 |
ENSDART00000184355
|
fam69ab
|
family with sequence similarity 69, member Ab |
| chr2_-_27619954 | 0.06 |
ENSDART00000144826
|
tgs1
|
trimethylguanosine synthase 1 |
| chr7_+_48297842 | 0.05 |
ENSDART00000052123
|
slc25a44b
|
solute carrier family 25, member 44 b |
| chr1_-_59139848 | 0.04 |
ENSDART00000191863
|
si:ch1073-110a20.2
|
si:ch1073-110a20.2 |
| chr2_-_12243213 | 0.04 |
ENSDART00000113081
|
gpr158b
|
G protein-coupled receptor 158b |
| chr6_+_23712911 | 0.03 |
ENSDART00000167795
|
zgc:158654
|
zgc:158654 |
| chr2_-_48966431 | 0.03 |
ENSDART00000147948
|
kcnj9
|
potassium inwardly-rectifying channel, subfamily J, member 9 |
| chr18_+_33276609 | 0.03 |
ENSDART00000137208
|
v2rx3
|
vomeronasal 2 receptor, x3 |
| chr13_+_406635 | 0.03 |
ENSDART00000181275
|
CU570800.4
|
|
| chr10_-_45058886 | 0.03 |
ENSDART00000159347
|
mrps24
|
mitochondrial ribosomal protein S24 |
| chr11_+_21587950 | 0.03 |
ENSDART00000162251
|
FAM72B
|
zgc:101564 |
| chr10_+_36345176 | 0.03 |
ENSDART00000099397
|
or105-1
|
odorant receptor, family C, subfamily 105, member 1 |
| chr2_+_6243144 | 0.02 |
ENSDART00000058258
|
gng5
|
guanine nucleotide binding protein (G protein), gamma 5 |
| chr24_+_14595680 | 0.02 |
ENSDART00000137337
ENSDART00000091784 ENSDART00000136026 |
thtpa
|
thiamine triphosphatase |
| chr15_-_931815 | 0.02 |
ENSDART00000106627
ENSDART00000102239 ENSDART00000184754 ENSDART00000186733 |
zgc:162936
|
zgc:162936 |
| chr6_-_17849786 | 0.02 |
ENSDART00000172709
|
rptor
|
regulatory associated protein of MTOR, complex 1 |
| chr21_-_22547496 | 0.01 |
ENSDART00000166835
ENSDART00000089030 |
myo5b
|
myosin VB |
| chr1_+_58535306 | 0.01 |
ENSDART00000165673
|
zgc:172091
|
zgc:172091 |
| chr4_-_75505048 | 0.00 |
ENSDART00000163665
|
si:dkey-71l4.5
|
si:dkey-71l4.5 |
| chr16_-_48071632 | 0.00 |
ENSDART00000148564
|
CSMD3 (1 of many)
|
si:ch211-236p22.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of pou6f1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) regulation of lamellipodium morphogenesis(GO:2000392) |
| 0.2 | 0.9 | GO:0019626 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) short-chain fatty acid catabolic process(GO:0019626) response to dexamethasone(GO:0071548) |
| 0.2 | 0.6 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 0.7 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.1 | 0.3 | GO:0060262 | N-terminal protein palmitoylation(GO:0006500) negative regulation of lipoprotein metabolic process(GO:0050748) regulation of N-terminal protein palmitoylation(GO:0060254) negative regulation of N-terminal protein palmitoylation(GO:0060262) negative regulation of protein lipidation(GO:1903060) |
| 0.1 | 0.3 | GO:0099543 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.1 | 0.6 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.1 | 0.3 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.1 | 0.4 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.1 | 0.3 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.1 | 1.3 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 1.2 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.4 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.4 | GO:1902868 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.0 | 0.8 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.6 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.2 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.0 | 0.1 | GO:0099612 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.0 | 0.7 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.0 | 3.3 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.7 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 1.0 | GO:0021854 | limbic system development(GO:0021761) hypothalamus development(GO:0021854) |
| 0.0 | 0.1 | GO:0032608 | interferon-beta production(GO:0032608) regulation of interferon-beta production(GO:0032648) positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.7 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.0 | 0.2 | GO:0031111 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.0 | 0.5 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.5 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.3 | GO:0006595 | polyamine metabolic process(GO:0006595) |
| 0.0 | 0.1 | GO:0030819 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.0 | 0.5 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 0.8 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.7 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.6 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 1.3 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.1 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.2 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.2 | GO:0031430 | M band(GO:0031430) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.2 | 0.6 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.1 | 0.7 | GO:0031843 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.1 | 1.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.7 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 0.4 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.1 | 0.7 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.3 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.8 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 0.6 | GO:0016832 | aldehyde-lyase activity(GO:0016832) oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 3.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.7 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.9 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.9 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.7 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 3.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.5 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.7 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.0 | 0.4 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.6 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |