Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
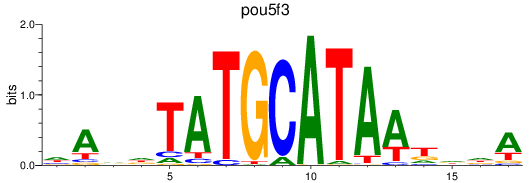
Results for pou5f3
Z-value: 0.15
Transcription factors associated with pou5f3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pou5f3
|
ENSDARG00000044774 | POU domain, class 5, transcription factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pou5f3 | dr11_v1_chr21_-_13690712_13690712 | -0.37 | 1.2e-01 | Click! |
Activity profile of pou5f3 motif
Sorted Z-values of pou5f3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_34868814 | 0.35 |
ENSDART00000153049
|
stmn4
|
stathmin-like 4 |
| chr22_+_15507218 | 0.33 |
ENSDART00000125450
|
gpc1a
|
glypican 1a |
| chr4_-_5302162 | 0.27 |
ENSDART00000177099
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr1_-_40994259 | 0.22 |
ENSDART00000101562
|
adra2c
|
adrenoceptor alpha 2C |
| chr22_+_5106751 | 0.17 |
ENSDART00000138967
|
atcaya
|
ataxia, cerebellar, Cayman type a |
| chr1_-_55068941 | 0.16 |
ENSDART00000152143
ENSDART00000152590 |
peli1a
|
pellino E3 ubiquitin protein ligase 1a |
| chr4_-_8903240 | 0.15 |
ENSDART00000129983
|
mpped1
|
metallophosphoesterase domain containing 1 |
| chr17_-_23609210 | 0.15 |
ENSDART00000064003
|
ifit15
|
interferon-induced protein with tetratricopeptide repeats 15 |
| chr7_-_72155022 | 0.12 |
ENSDART00000187984
|
zmp:0000001168
|
zmp:0000001168 |
| chr7_+_61050329 | 0.12 |
ENSDART00000115355
|
nwd2
|
NACHT and WD repeat domain containing 2 |
| chr5_-_30074332 | 0.11 |
ENSDART00000147963
|
bco2a
|
beta-carotene oxygenase 2a |
| chr18_-_8030073 | 0.11 |
ENSDART00000151490
|
shank3a
|
SH3 and multiple ankyrin repeat domains 3a |
| chr9_-_2945008 | 0.10 |
ENSDART00000183452
|
zak
|
sterile alpha motif and leucine zipper containing kinase AZK |
| chr13_-_39830999 | 0.09 |
ENSDART00000115089
|
zgc:171482
|
zgc:171482 |
| chr10_+_20628698 | 0.08 |
ENSDART00000064666
|
prnpb
|
prion protein b |
| chr24_-_25004553 | 0.08 |
ENSDART00000080997
ENSDART00000136860 |
zdhhc20b
|
zinc finger, DHHC-type containing 20b |
| chr24_-_7321928 | 0.07 |
ENSDART00000167570
ENSDART00000045150 |
actr3b
|
ARP3 actin related protein 3 homolog B |
| chr11_-_36001495 | 0.07 |
ENSDART00000190330
|
itpr1b
|
inositol 1,4,5-trisphosphate receptor, type 1b |
| chr25_+_26923193 | 0.07 |
ENSDART00000187364
|
grm8b
|
glutamate receptor, metabotropic 8b |
| chr24_+_3328354 | 0.07 |
ENSDART00000147468
|
bdh1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr24_+_17007407 | 0.06 |
ENSDART00000110652
|
zfx
|
zinc finger protein, X-linked |
| chr13_+_31172833 | 0.06 |
ENSDART00000176378
|
CR931802.3
|
|
| chr18_-_29925717 | 0.05 |
ENSDART00000099281
|
mhc2dbb
|
major histocompatibility complex class II DBB gene |
| chr8_+_7740004 | 0.05 |
ENSDART00000170184
ENSDART00000187811 |
fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr21_+_39361040 | 0.05 |
ENSDART00000085453
|
cluhb
|
clustered mitochondria (cluA/CLU1) homolog b |
| chr8_-_22698651 | 0.05 |
ENSDART00000181411
|
iqsec2a
|
IQ motif and Sec7 domain 2a |
| chr19_-_43552252 | 0.05 |
ENSDART00000138308
|
gpr186
|
G protein-coupled receptor 186 |
| chr1_+_58442694 | 0.05 |
ENSDART00000160897
|
zgc:194906
|
zgc:194906 |
| chr16_-_31351419 | 0.04 |
ENSDART00000178298
ENSDART00000018091 |
mroh1
|
maestro heat-like repeat family member 1 |
| chr2_-_20120904 | 0.04 |
ENSDART00000186002
ENSDART00000124724 |
dpydb
|
dihydropyrimidine dehydrogenase b |
| chr9_+_12444494 | 0.04 |
ENSDART00000102430
|
tmem41aa
|
transmembrane protein 41aa |
| chr11_+_19060079 | 0.04 |
ENSDART00000158123
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr18_+_15758375 | 0.02 |
ENSDART00000137554
|
si:ch211-219a15.4
|
si:ch211-219a15.4 |
| chr16_-_42523744 | 0.02 |
ENSDART00000017185
|
tbx20
|
T-box 20 |
| chr22_-_30881738 | 0.01 |
ENSDART00000059965
|
si:dkey-49n23.1
|
si:dkey-49n23.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of pou5f3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.0 | 0.2 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.0 | 0.1 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.1 | GO:0098815 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.0 | 0.2 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 0.0 | GO:0046104 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.0 | 0.1 | GO:0042214 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.0 | 0.3 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.2 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 0.3 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.1 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |