Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
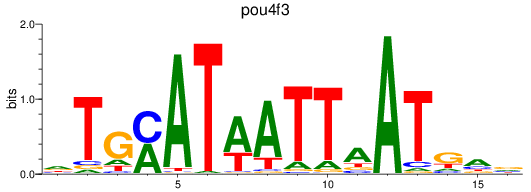
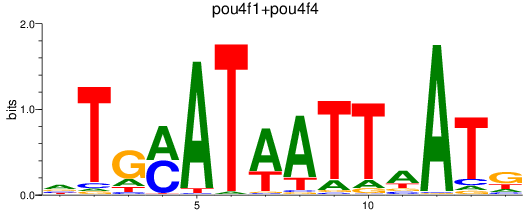
Results for pou4f3_pou4f1+pou4f4
Z-value: 1.62
Transcription factors associated with pou4f3_pou4f1+pou4f4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pou4f3
|
ENSDARG00000006206 | POU class 4 homeobox 3 |
|
pou4f1
|
ENSDARG00000005559 | POU class 4 homeobox 1 |
|
pou4f4
|
ENSDARG00000044375 | zgc |
|
pou4f4
|
ENSDARG00000111929 | zgc |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pou4f3 | dr11_v1_chr9_+_54290896_54290896 | 0.78 | 9.0e-05 | Click! |
| pou4f1 | dr11_v1_chr6_+_4539953_4539953 | 0.25 | 3.0e-01 | Click! |
| zgc:158291 | dr11_v1_chr21_+_38312549_38312549 | -0.03 | 9.1e-01 | Click! |
Activity profile of pou4f3_pou4f1+pou4f4 motif
Sorted Z-values of pou4f3_pou4f1+pou4f4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_30620625 | 8.05 |
ENSDART00000098273
|
tcnl
|
transcobalamin like |
| chr11_+_6819050 | 7.64 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr19_-_10196370 | 6.90 |
ENSDART00000091707
|
dbpa
|
D site albumin promoter binding protein a |
| chr17_+_2162916 | 5.88 |
ENSDART00000103775
|
pak6a
|
p21 protein (Cdc42/Rac)-activated kinase 6a |
| chr7_-_69298271 | 5.73 |
ENSDART00000122935
|
phlpp2
|
PH domain and leucine rich repeat protein phosphatase 2 |
| chr1_-_21901589 | 5.29 |
ENSDART00000140553
|
frmpd1a
|
FERM and PDZ domain containing 1a |
| chr20_-_40754794 | 5.10 |
ENSDART00000187251
|
cx32.3
|
connexin 32.3 |
| chr2_+_39108339 | 5.00 |
ENSDART00000085675
|
clstn2
|
calsyntenin 2 |
| chr5_+_51079504 | 4.95 |
ENSDART00000097466
|
fam169aa
|
family with sequence similarity 169, member Aa |
| chr23_-_27345425 | 4.93 |
ENSDART00000022042
ENSDART00000191870 |
scn8aa
|
sodium channel, voltage gated, type VIII, alpha subunit a |
| chr5_-_30418636 | 4.67 |
ENSDART00000098285
|
atf5a
|
activating transcription factor 5a |
| chr25_-_4148719 | 4.58 |
ENSDART00000112880
ENSDART00000023278 |
fads2
|
fatty acid desaturase 2 |
| chr24_-_4765740 | 4.53 |
ENSDART00000121576
|
cpb1
|
carboxypeptidase B1 (tissue) |
| chr25_-_13614702 | 4.47 |
ENSDART00000165510
ENSDART00000190959 |
fa2h
|
fatty acid 2-hydroxylase |
| chr7_-_34225011 | 4.45 |
ENSDART00000049588
|
crybgx
|
crystallin beta gamma X |
| chr20_+_9355912 | 4.38 |
ENSDART00000142337
|
si:dkey-174m14.3
|
si:dkey-174m14.3 |
| chr5_-_23429228 | 4.22 |
ENSDART00000049291
|
gria3a
|
glutamate receptor, ionotropic, AMPA 3a |
| chr4_+_12111154 | 4.07 |
ENSDART00000036779
|
tmem178b
|
transmembrane protein 178B |
| chr16_+_14029283 | 3.89 |
ENSDART00000146165
ENSDART00000132075 |
rusc1
|
RUN and SH3 domain containing 1 |
| chr20_-_31808779 | 3.85 |
ENSDART00000133788
|
stxbp5a
|
syntaxin binding protein 5a (tomosyn) |
| chr6_-_39489190 | 3.85 |
ENSDART00000151299
|
scn8ab
|
sodium channel, voltage gated, type VIII, alpha subunit b |
| chr25_+_28679672 | 3.57 |
ENSDART00000139965
ENSDART00000134072 |
cnot2
|
CCR4-NOT transcription complex, subunit 2 |
| chr16_-_49505275 | 3.29 |
ENSDART00000160784
|
satb1b
|
SATB homeobox 1b |
| chr5_+_58372164 | 3.28 |
ENSDART00000057910
|
nrgna
|
neurogranin (protein kinase C substrate, RC3) a |
| chr16_-_12953739 | 3.27 |
ENSDART00000103894
|
cacng8b
|
calcium channel, voltage-dependent, gamma subunit 8b |
| chr6_+_21740672 | 3.26 |
ENSDART00000193734
|
lhfpl4a
|
lipoma HMGIC fusion partner-like 4a |
| chr13_-_43599898 | 3.24 |
ENSDART00000084416
ENSDART00000145705 |
ablim1a
|
actin binding LIM protein 1a |
| chr6_+_45494227 | 3.18 |
ENSDART00000159863
|
cntn4
|
contactin 4 |
| chr2_+_25929619 | 3.15 |
ENSDART00000137746
|
slc7a14a
|
solute carrier family 7, member 14a |
| chr10_+_38775408 | 3.10 |
ENSDART00000125045
|
dscama
|
Down syndrome cell adhesion molecule a |
| chr9_-_22213297 | 3.04 |
ENSDART00000110656
ENSDART00000133149 |
crygm2d20
|
crystallin, gamma M2d20 |
| chr17_-_37395460 | 3.00 |
ENSDART00000148160
ENSDART00000075975 |
crip1
|
cysteine-rich protein 1 |
| chr20_+_22045089 | 3.00 |
ENSDART00000063564
ENSDART00000187013 ENSDART00000161552 ENSDART00000174478 ENSDART00000063568 ENSDART00000152247 |
nmu
|
neuromedin U |
| chr22_+_5687615 | 2.98 |
ENSDART00000133241
ENSDART00000019854 ENSDART00000138102 |
dnase1l4.2
|
deoxyribonuclease 1 like 4, tandem duplicate 2 |
| chr2_-_43196595 | 2.93 |
ENSDART00000141087
|
crema
|
cAMP responsive element modulator a |
| chr20_-_44575103 | 2.85 |
ENSDART00000192573
|
ubxn2a
|
UBX domain protein 2A |
| chr16_+_16849220 | 2.84 |
ENSDART00000047409
ENSDART00000142155 |
myh14
|
myosin, heavy chain 14, non-muscle |
| chr21_-_13123176 | 2.77 |
ENSDART00000144866
ENSDART00000024616 |
fam219aa
|
family with sequence similarity 219, member Aa |
| chr23_-_29376859 | 2.71 |
ENSDART00000146411
|
sst6
|
somatostatin 6 |
| chr20_-_34801181 | 2.67 |
ENSDART00000048375
ENSDART00000132426 |
stmn4
|
stathmin-like 4 |
| chr23_-_24195723 | 2.65 |
ENSDART00000145489
|
ano11
|
anoctamin 11 |
| chr1_-_15797663 | 2.64 |
ENSDART00000177122
|
SGCZ
|
sarcoglycan zeta |
| chr22_+_15507218 | 2.59 |
ENSDART00000125450
|
gpc1a
|
glypican 1a |
| chr16_+_10776688 | 2.58 |
ENSDART00000161969
ENSDART00000172657 |
atp1a3b
|
ATPase Na+/K+ transporting subunit alpha 3b |
| chr22_-_2959005 | 2.58 |
ENSDART00000040701
|
ing5a
|
inhibitor of growth family, member 5a |
| chr6_-_35446110 | 2.56 |
ENSDART00000058773
|
rgs16
|
regulator of G protein signaling 16 |
| chr25_+_24291156 | 2.51 |
ENSDART00000083407
|
b4galnt4a
|
beta-1,4-N-acetyl-galactosaminyl transferase 4a |
| chr11_+_23760470 | 2.39 |
ENSDART00000175688
ENSDART00000121874 ENSDART00000086720 |
nfasca
|
neurofascin homolog (chicken) a |
| chr25_-_13614863 | 2.36 |
ENSDART00000121859
|
fa2h
|
fatty acid 2-hydroxylase |
| chr14_-_18672561 | 2.35 |
ENSDART00000166730
ENSDART00000006998 |
slitrk4
|
SLIT and NTRK-like family, member 4 |
| chr5_-_30074332 | 2.34 |
ENSDART00000147963
|
bco2a
|
beta-carotene oxygenase 2a |
| chr19_-_3574060 | 2.33 |
ENSDART00000105120
|
tmem170b
|
transmembrane protein 170B |
| chr16_-_45398408 | 2.31 |
ENSDART00000004052
|
rgl2
|
ral guanine nucleotide dissociation stimulator-like 2 |
| chr11_-_38083397 | 2.27 |
ENSDART00000086516
ENSDART00000184033 |
klhdc8a
|
kelch domain containing 8A |
| chr10_-_10018120 | 2.16 |
ENSDART00000132375
|
strbp
|
spermatid perinuclear RNA binding protein |
| chr9_+_38481780 | 2.15 |
ENSDART00000087241
|
lss
|
lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) |
| chr10_+_38775959 | 2.14 |
ENSDART00000192990
|
dscama
|
Down syndrome cell adhesion molecule a |
| chr21_+_27278120 | 2.13 |
ENSDART00000193882
|
si:dkey-175m17.7
|
si:dkey-175m17.7 |
| chr13_+_28702104 | 2.09 |
ENSDART00000135481
|
si:ch211-67n3.9
|
si:ch211-67n3.9 |
| chr10_+_25726694 | 2.08 |
ENSDART00000140308
|
ugt5d1
|
UDP glucuronosyltransferase 5 family, polypeptide D1 |
| chr8_-_22838364 | 1.98 |
ENSDART00000187954
ENSDART00000185368 ENSDART00000188210 |
magixa
|
MAGI family member, X-linked a |
| chr12_+_2446837 | 1.97 |
ENSDART00000112032
|
ARHGAP22
|
si:dkey-191m6.4 |
| chr11_+_37137196 | 1.86 |
ENSDART00000172950
|
wnk2
|
WNK lysine deficient protein kinase 2 |
| chr13_+_45524475 | 1.83 |
ENSDART00000074567
ENSDART00000019113 |
maco1b
|
macoilin 1b |
| chr8_-_20230559 | 1.82 |
ENSDART00000193677
|
mllt1a
|
MLLT1, super elongation complex subunit a |
| chr17_-_20287530 | 1.81 |
ENSDART00000078703
ENSDART00000191289 |
add3b
|
adducin 3 (gamma) b |
| chr24_-_26622423 | 1.78 |
ENSDART00000182044
|
tnikb
|
TRAF2 and NCK interacting kinase b |
| chr5_-_68058168 | 1.76 |
ENSDART00000177026
|
rnf167
|
ring finger protein 167 |
| chr2_+_50608099 | 1.75 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr20_+_41664350 | 1.74 |
ENSDART00000186247
|
fam184a
|
family with sequence similarity 184, member A |
| chr8_+_7975745 | 1.74 |
ENSDART00000137920
|
si:ch211-169p10.1
|
si:ch211-169p10.1 |
| chr23_-_24195519 | 1.72 |
ENSDART00000112370
ENSDART00000180377 |
ano11
|
anoctamin 11 |
| chr18_+_15616167 | 1.72 |
ENSDART00000080454
|
slc17a8
|
solute carrier family 17 (vesicular glutamate transporter), member 8 |
| chr13_-_17860307 | 1.71 |
ENSDART00000135920
ENSDART00000054579 |
march8
|
membrane-associated ring finger (C3HC4) 8 |
| chr13_-_49666615 | 1.67 |
ENSDART00000148083
|
tomm20a
|
translocase of outer mitochondrial membrane 20 |
| chr24_+_14713776 | 1.64 |
ENSDART00000134475
|
gdap1
|
ganglioside induced differentiation associated protein 1 |
| chr2_+_22702488 | 1.62 |
ENSDART00000076647
|
kif1ab
|
kinesin family member 1Ab |
| chr7_+_73397283 | 1.60 |
ENSDART00000174390
|
CABZ01081780.1
|
|
| chr11_-_38513978 | 1.59 |
ENSDART00000123381
|
si:ch211-117k10.3
|
si:ch211-117k10.3 |
| chr15_-_14552101 | 1.59 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr3_-_23407720 | 1.57 |
ENSDART00000155658
|
rapgefl1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr12_-_26406323 | 1.55 |
ENSDART00000131896
|
myoz1b
|
myozenin 1b |
| chr2_-_31651297 | 1.55 |
ENSDART00000132955
|
si:ch211-106h4.9
|
si:ch211-106h4.9 |
| chr23_-_18024543 | 1.54 |
ENSDART00000139695
|
pm20d1.1
|
peptidase M20 domain containing 1, tandem duplicate 1 |
| chr18_-_47662696 | 1.53 |
ENSDART00000184260
|
CABZ01073963.1
|
|
| chr16_-_38118003 | 1.52 |
ENSDART00000058667
|
si:dkey-23o4.6
|
si:dkey-23o4.6 |
| chr23_+_6232895 | 1.50 |
ENSDART00000139795
|
syt2a
|
synaptotagmin IIa |
| chr7_-_71829649 | 1.50 |
ENSDART00000160449
|
cacnb2a
|
calcium channel, voltage-dependent, beta 2a |
| chr3_+_26734162 | 1.44 |
ENSDART00000114552
|
si:dkey-202l16.5
|
si:dkey-202l16.5 |
| chr20_+_31076488 | 1.43 |
ENSDART00000136255
ENSDART00000008840 |
otofa
|
otoferlin a |
| chr23_+_20689255 | 1.42 |
ENSDART00000182420
|
usp21
|
ubiquitin specific peptidase 21 |
| chr20_-_48172556 | 1.41 |
ENSDART00000097888
|
CABZ01059099.1
|
|
| chr17_-_51893123 | 1.40 |
ENSDART00000103350
ENSDART00000017329 |
numb
|
numb homolog (Drosophila) |
| chr13_+_8604710 | 1.36 |
ENSDART00000091097
|
socs5b
|
suppressor of cytokine signaling 5b |
| chr6_+_38427570 | 1.34 |
ENSDART00000170612
|
gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr23_-_27692717 | 1.33 |
ENSDART00000053878
ENSDART00000145028 |
IKZF4
|
si:dkey-166n8.9 |
| chr11_-_1956204 | 1.32 |
ENSDART00000185541
|
nr1d4b
|
nuclear receptor subfamily 1, group D, member 4b |
| chr3_-_28048475 | 1.31 |
ENSDART00000150888
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr17_+_31914877 | 1.30 |
ENSDART00000177801
|
FAM196A (1 of many)
|
family with sequence similarity 196 member A |
| chr24_-_3419998 | 1.28 |
ENSDART00000066839
|
slc35g2b
|
solute carrier family 35, member G2b |
| chr7_-_49892991 | 1.27 |
ENSDART00000126240
|
cd44a
|
CD44 molecule (Indian blood group) a |
| chr9_-_68934 | 1.25 |
ENSDART00000054594
ENSDART00000009389 |
il10rb
|
interleukin 10 receptor, beta |
| chr12_-_26537145 | 1.24 |
ENSDART00000138437
ENSDART00000163931 ENSDART00000132737 |
acsf2
|
acyl-CoA synthetase family member 2 |
| chr17_+_15983247 | 1.24 |
ENSDART00000154275
|
clmn
|
calmin |
| chr1_+_44491077 | 1.24 |
ENSDART00000073736
|
rtn4rl2a
|
reticulon 4 receptor-like 2 a |
| chr11_+_44356504 | 1.23 |
ENSDART00000160678
|
srsf7b
|
serine/arginine-rich splicing factor 7b |
| chr8_+_668184 | 1.22 |
ENSDART00000183788
|
rnf165b
|
ring finger protein 165b |
| chr13_-_21688176 | 1.22 |
ENSDART00000063825
|
sprn
|
shadow of prion protein |
| chr3_-_61336841 | 1.21 |
ENSDART00000155414
|
tecpr1b
|
tectonin beta-propeller repeat containing 1b |
| chr23_-_30431333 | 1.20 |
ENSDART00000146633
|
camta1a
|
calmodulin binding transcription activator 1a |
| chr24_+_3328354 | 1.20 |
ENSDART00000147468
|
bdh1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr6_+_4299164 | 1.18 |
ENSDART00000159759
|
nbeal1
|
neurobeachin-like 1 |
| chr23_+_21966447 | 1.18 |
ENSDART00000189378
|
lactbl1a
|
lactamase, beta-like 1a |
| chr10_+_42423318 | 1.17 |
ENSDART00000134282
|
npy8ar
|
neuropeptide Y receptor Y8a |
| chr13_+_12299997 | 1.16 |
ENSDART00000108535
|
gabrb1
|
gamma-aminobutyric acid (GABA) A receptor, beta 1 |
| chr6_+_3280939 | 1.14 |
ENSDART00000151359
|
kdm4aa
|
lysine (K)-specific demethylase 4A, genome duplicate a |
| chr9_+_22388686 | 1.13 |
ENSDART00000182731
ENSDART00000181462 |
dgkg
|
diacylglycerol kinase, gamma |
| chr3_-_23406964 | 1.11 |
ENSDART00000114723
|
rapgefl1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr1_+_36665555 | 1.10 |
ENSDART00000128557
ENSDART00000010632 |
ednraa
|
endothelin receptor type Aa |
| chr9_-_18735256 | 1.10 |
ENSDART00000143165
|
tsc22d1
|
TSC22 domain family, member 1 |
| chr7_+_27976448 | 1.09 |
ENSDART00000181026
|
tub
|
tubby bipartite transcription factor |
| chr2_-_48171112 | 1.09 |
ENSDART00000156258
|
pfkpb
|
phosphofructokinase, platelet b |
| chr15_+_45586471 | 1.09 |
ENSDART00000165699
|
cldn15lb
|
claudin 15-like b |
| chr11_+_19060079 | 1.09 |
ENSDART00000158123
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr16_+_29630965 | 1.06 |
ENSDART00000185820
ENSDART00000192105 ENSDART00000153683 ENSDART00000186713 |
VPS72
|
si:ch211-203d17.1 |
| chr2_-_50966124 | 1.05 |
ENSDART00000170470
|
ghrhra
|
growth hormone releasing hormone receptor a |
| chr13_+_31757331 | 1.05 |
ENSDART00000044282
|
hif1aa
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) a |
| chr6_-_41229787 | 1.05 |
ENSDART00000065013
|
synpr
|
synaptoporin |
| chr9_-_40931637 | 1.05 |
ENSDART00000165103
|
C2orf88
|
Small membrane A-kinase anchor protein |
| chr11_+_23957440 | 1.04 |
ENSDART00000190721
|
cntn2
|
contactin 2 |
| chr3_-_34717882 | 1.03 |
ENSDART00000151127
|
thraa
|
thyroid hormone receptor alpha a |
| chr3_-_28665291 | 1.02 |
ENSDART00000151670
|
fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr7_-_12371206 | 1.01 |
ENSDART00000161679
|
efl1
|
elongation factor like GTPase 1 |
| chr13_-_32635859 | 1.00 |
ENSDART00000146249
ENSDART00000145395 ENSDART00000148040 ENSDART00000100650 |
matn3b
|
matrilin 3b |
| chr4_+_6869847 | 0.99 |
ENSDART00000036646
|
dock4b
|
dedicator of cytokinesis 4b |
| chr11_-_45135643 | 0.98 |
ENSDART00000170863
|
cant1b
|
calcium activated nucleotidase 1b |
| chr11_+_44356707 | 0.96 |
ENSDART00000165219
|
srsf7b
|
serine/arginine-rich splicing factor 7b |
| chr20_-_2361226 | 0.95 |
ENSDART00000172130
|
EPB41L2
|
si:ch73-18b11.1 |
| chr9_+_11034967 | 0.95 |
ENSDART00000152081
|
asic4a
|
acid-sensing (proton-gated) ion channel family member 4a |
| chr7_+_3357973 | 0.94 |
ENSDART00000172853
|
si:ch211-285c6.5
|
si:ch211-285c6.5 |
| chr15_+_32297441 | 0.94 |
ENSDART00000153657
|
trim3a
|
tripartite motif containing 3a |
| chr6_+_8079974 | 0.93 |
ENSDART00000152071
|
si:ch211-207j7.2
|
si:ch211-207j7.2 |
| chr7_-_28148310 | 0.91 |
ENSDART00000044208
|
lmo1
|
LIM domain only 1 |
| chr10_+_40214877 | 0.89 |
ENSDART00000109689
|
gramd1ba
|
GRAM domain containing 1Ba |
| chr13_-_39830999 | 0.87 |
ENSDART00000115089
|
zgc:171482
|
zgc:171482 |
| chr23_+_33907899 | 0.85 |
ENSDART00000159445
|
cs
|
citrate synthase |
| chr10_+_19528321 | 0.85 |
ENSDART00000184816
|
vsig8a
|
V-set and immunoglobulin domain containing 8a |
| chr17_+_33999630 | 0.83 |
ENSDART00000167085
ENSDART00000155030 ENSDART00000168522 ENSDART00000191799 ENSDART00000189684 ENSDART00000153942 ENSDART00000187272 ENSDART00000127692 |
gphna
|
gephyrin a |
| chr22_-_32360464 | 0.83 |
ENSDART00000148886
|
pcbp4
|
poly(rC) binding protein 4 |
| chr20_+_27298783 | 0.83 |
ENSDART00000013861
|
ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chr9_+_7030016 | 0.82 |
ENSDART00000148047
ENSDART00000148181 |
inpp4aa
|
inositol polyphosphate-4-phosphatase type I Aa |
| chr1_+_31113951 | 0.82 |
ENSDART00000129362
|
eef1a1b
|
eukaryotic translation elongation factor 1 alpha 1b |
| chr25_+_8407892 | 0.81 |
ENSDART00000153536
|
muc5.2
|
mucin 5.2 |
| chr17_+_15983557 | 0.81 |
ENSDART00000190806
|
clmn
|
calmin |
| chr12_-_30523216 | 0.81 |
ENSDART00000152896
ENSDART00000153191 |
plekhs1
|
pleckstrin homology domain containing, family S member 1 |
| chr14_-_7207961 | 0.80 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr10_+_5645887 | 0.79 |
ENSDART00000171426
|
pdzph1
|
PDZ and pleckstrin homology domains 1 |
| chr20_-_43663494 | 0.79 |
ENSDART00000144564
|
BX470188.1
|
|
| chr7_+_53156810 | 0.78 |
ENSDART00000189816
|
cdh29
|
cadherin 29 |
| chr1_-_43899422 | 0.77 |
ENSDART00000134649
|
si:dkey-22i16.2
|
si:dkey-22i16.2 |
| chr5_+_4054704 | 0.77 |
ENSDART00000140537
|
dhrs11a
|
dehydrogenase/reductase (SDR family) member 11a |
| chr22_-_20379045 | 0.76 |
ENSDART00000183511
|
zbtb7a
|
zinc finger and BTB domain containing 7a |
| chr8_-_27858458 | 0.76 |
ENSDART00000132632
ENSDART00000136562 |
cttnbp2nlb
|
CTTNBP2 N-terminal like b |
| chr7_+_47316944 | 0.76 |
ENSDART00000173534
|
dpy19l3
|
dpy-19 like C-mannosyltransferase 3 |
| chr12_+_20618336 | 0.75 |
ENSDART00000084007
|
si:dkey-220f10.4
|
si:dkey-220f10.4 |
| chr18_+_40355408 | 0.75 |
ENSDART00000167134
|
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr13_+_36622100 | 0.73 |
ENSDART00000133198
|
si:ch211-67f24.7
|
si:ch211-67f24.7 |
| chr21_+_170038 | 0.69 |
ENSDART00000157614
|
klhl8
|
kelch-like family member 8 |
| chr11_+_36409457 | 0.68 |
ENSDART00000077641
|
cyb561d1
|
cytochrome b561 family, member D1 |
| chr12_-_33893381 | 0.67 |
ENSDART00000153280
|
sema4gb
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Gb |
| chr1_+_25783801 | 0.66 |
ENSDART00000102455
|
gucy1a1
|
guanylate cyclase 1 soluble subunit alpha 1 |
| chr19_-_43552252 | 0.66 |
ENSDART00000138308
|
gpr186
|
G protein-coupled receptor 186 |
| chr20_-_39333657 | 0.66 |
ENSDART00000153720
ENSDART00000142164 |
ccl38.1
|
chemokine (C-C motif) ligand 38, duplicate 1 |
| chr3_-_30434016 | 0.65 |
ENSDART00000150958
|
lrrc4ba
|
leucine rich repeat containing 4Ba |
| chr11_+_34523132 | 0.64 |
ENSDART00000192257
|
zmat3
|
zinc finger, matrin-type 3 |
| chr22_-_14475927 | 0.64 |
ENSDART00000135768
|
lrp1ba
|
low density lipoprotein receptor-related protein 1Ba |
| chr21_+_5882300 | 0.63 |
ENSDART00000165065
|
uqcr10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr8_-_39952727 | 0.61 |
ENSDART00000181310
|
cabp1a
|
calcium binding protein 1a |
| chr24_+_36840652 | 0.61 |
ENSDART00000088168
|
CABZ01055347.1
|
|
| chr3_+_22578369 | 0.61 |
ENSDART00000187695
ENSDART00000182678 ENSDART00000112270 |
tanc2a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2a |
| chr16_-_31351419 | 0.60 |
ENSDART00000178298
ENSDART00000018091 |
mroh1
|
maestro heat-like repeat family member 1 |
| chr1_+_38758261 | 0.60 |
ENSDART00000182756
|
wdr17
|
WD repeat domain 17 |
| chr2_-_9818640 | 0.59 |
ENSDART00000139499
ENSDART00000165548 ENSDART00000012442 ENSDART00000046587 |
ap2m1b
|
adaptor-related protein complex 2, mu 1 subunit, b |
| chr8_-_23884301 | 0.57 |
ENSDART00000185509
ENSDART00000147202 |
lhfpl5b
|
LHFPL tetraspan subfamily member 5b |
| chr23_+_41831224 | 0.56 |
ENSDART00000171885
|
scp2b
|
sterol carrier protein 2b |
| chr13_-_16066997 | 0.54 |
ENSDART00000184790
|
SPATA48
|
spermatogenesis associated 48 |
| chr12_+_20618522 | 0.54 |
ENSDART00000091452
|
si:dkey-220f10.4
|
si:dkey-220f10.4 |
| chr7_+_27317174 | 0.53 |
ENSDART00000193058
|
sox6
|
SRY (sex determining region Y)-box 6 |
| chr2_+_45511459 | 0.52 |
ENSDART00000132101
|
aknad1
|
AKNA domain containing 1 |
| chr4_+_9669717 | 0.52 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr16_+_1353894 | 0.51 |
ENSDART00000148426
|
celf3b
|
cugbp, Elav-like family member 3b |
| chr11_+_30057762 | 0.51 |
ENSDART00000164139
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr10_+_44824072 | 0.46 |
ENSDART00000162669
|
slc4a5
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 5 |
| chr9_-_10532591 | 0.46 |
ENSDART00000175269
|
thsd7ba
|
thrombospondin, type I, domain containing 7Ba |
| chr1_+_38758445 | 0.46 |
ENSDART00000136300
|
wdr17
|
WD repeat domain 17 |
| chr20_-_40766387 | 0.45 |
ENSDART00000061173
|
hsdl1
|
hydroxysteroid dehydrogenase like 1 |
| chr11_-_25777318 | 0.45 |
ENSDART00000079328
|
plch2b
|
phospholipase C, eta 2b |
| chr22_+_30517227 | 0.44 |
ENSDART00000170978
|
si:dkey-103k4.1
|
si:dkey-103k4.1 |
| chr20_-_11178022 | 0.43 |
ENSDART00000152246
|
flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr7_+_6941583 | 0.43 |
ENSDART00000160709
ENSDART00000157634 |
rbm14b
|
RNA binding motif protein 14b |
Network of associatons between targets according to the STRING database.
First level regulatory network of pou4f3_pou4f1+pou4f4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.6 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 1.2 | 4.9 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 1.0 | 4.1 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 1.0 | 3.0 | GO:0097237 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.4 | 4.4 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.4 | 1.7 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.4 | 2.1 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.4 | 1.5 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.4 | 1.8 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.4 | 3.6 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.3 | 1.0 | GO:0048677 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.3 | 1.0 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.3 | 1.1 | GO:0010155 | regulation of proton transport(GO:0010155) |
| 0.3 | 2.3 | GO:0046247 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.3 | 4.4 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.3 | 0.8 | GO:0018211 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.2 | 3.8 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.2 | 3.0 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.2 | 1.7 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.2 | 3.3 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.2 | 5.2 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.2 | 3.0 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.2 | 2.9 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.2 | 0.8 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.1 | 3.2 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 3.7 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.1 | 2.6 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 2.6 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.1 | 1.4 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.1 | 0.3 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.1 | 1.9 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.1 | 0.6 | GO:0035678 | neuromast hair cell morphogenesis(GO:0035678) |
| 0.1 | 0.6 | GO:0032370 | positive regulation of lipid transport(GO:0032370) |
| 0.1 | 0.8 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 1.0 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.1 | 3.3 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.1 | 1.6 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 1.1 | GO:0061718 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 3.3 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.1 | 3.9 | GO:0050708 | regulation of protein secretion(GO:0050708) |
| 0.1 | 10.8 | GO:0006633 | fatty acid biosynthetic process(GO:0006633) |
| 0.1 | 0.8 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.1 | 0.7 | GO:0090594 | inflammatory response to wounding(GO:0090594) |
| 0.1 | 1.2 | GO:0048922 | posterior lateral line neuromast deposition(GO:0048922) |
| 0.1 | 2.1 | GO:0016114 | terpenoid biosynthetic process(GO:0016114) |
| 0.1 | 1.5 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.2 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.1 | 1.4 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.1 | 0.6 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 8.0 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.1 | 0.2 | GO:0070257 | regulation of mucus secretion(GO:0070255) positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 2.7 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 2.4 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.1 | 0.9 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.2 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.0 | 1.1 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 4.5 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 1.1 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.0 | 0.2 | GO:1901380 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 1.8 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.3 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.4 | GO:0006566 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.0 | 0.4 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.0 | 1.5 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 1.4 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 1.3 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 3.2 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.0 | 0.9 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 1.4 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 1.7 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 2.2 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 0.2 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 1.7 | GO:0042552 | myelination(GO:0042552) |
| 0.0 | 1.9 | GO:0070374 | positive regulation of ERK1 and ERK2 cascade(GO:0070374) |
| 0.0 | 0.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.5 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 1.0 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.3 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.9 | GO:0009755 | hormone-mediated signaling pathway(GO:0009755) |
| 0.0 | 1.9 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.0 | 2.4 | GO:0030334 | regulation of cell migration(GO:0030334) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.4 | 8.8 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.3 | 1.5 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.3 | 3.6 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.2 | 7.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.2 | 1.7 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 2.6 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 1.6 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 1.7 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 2.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 1.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 1.0 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 1.8 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 2.5 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.6 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 3.2 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 4.0 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 1.4 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.6 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 8.7 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 1.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 1.5 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 1.9 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 2.8 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 1.7 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 6.4 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 1.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.6 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.8 | GO:1903293 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 8.7 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 1.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.4 | GO:0016607 | nuclear speck(GO:0016607) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 6.8 | GO:0080132 | fatty acid alpha-hydroxylase activity(GO:0080132) |
| 0.5 | 2.5 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.4 | 1.2 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.4 | 9.9 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.4 | 1.2 | GO:0031956 | medium-chain fatty acid-CoA ligase activity(GO:0031956) butyrate-CoA ligase activity(GO:0047760) |
| 0.3 | 3.0 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.3 | 4.2 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.3 | 0.8 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.3 | 2.3 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.3 | 2.6 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.2 | 3.9 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.2 | 4.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 5.2 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.2 | 1.1 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.2 | 3.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.2 | 1.6 | GO:0051373 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.2 | 1.5 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.2 | 2.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 1.7 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.1 | 1.4 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.8 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.1 | 1.9 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 2.6 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 0.9 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 1.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 0.4 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.1 | 7.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 1.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.4 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.1 | 2.5 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 3.0 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 0.2 | GO:0033961 | cis-stilbene-oxide hydrolase activity(GO:0033961) |
| 0.1 | 2.4 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 1.2 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.1 | 0.4 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.3 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 4.8 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 1.5 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 1.7 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 1.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 1.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 1.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 2.6 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 2.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 1.2 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 1.1 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 1.0 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.7 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.4 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 2.2 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.3 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 2.9 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 3.2 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.3 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 4.4 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 2.6 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 3.3 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.6 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.8 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 9.1 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 1.0 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 3.3 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.2 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.5 | GO:0016229 | steroid dehydrogenase activity(GO:0016229) |
| 0.0 | 2.3 | GO:0042277 | peptide binding(GO:0042277) |
| 0.0 | 5.1 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.6 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 4.3 | GO:0003924 | GTPase activity(GO:0003924) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.9 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 1.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.4 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.4 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.2 | PID EPHA FWDPATHWAY | EPHA forward signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.6 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.2 | 3.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.2 | 2.8 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.2 | 2.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 2.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 0.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.4 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.9 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 1.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 3.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.0 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 2.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |