Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
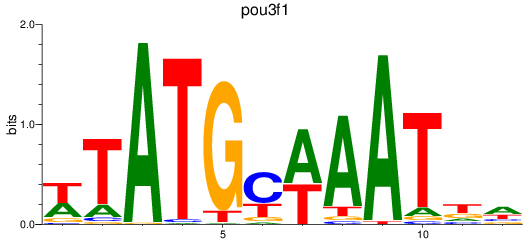
Results for pou3f1
Z-value: 0.92
Transcription factors associated with pou3f1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pou3f1
|
ENSDARG00000009823 | POU class 3 homeobox 1 |
|
pou3f1
|
ENSDARG00000112302 | POU class 3 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pou3f1 | dr11_v1_chr16_+_33593116_33593116 | -0.39 | 1.0e-01 | Click! |
Activity profile of pou3f1 motif
Sorted Z-values of pou3f1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_30563115 | 3.13 |
ENSDART00000028566
|
si:ch211-200p22.4
|
si:ch211-200p22.4 |
| chr13_-_7031033 | 2.84 |
ENSDART00000193211
|
CABZ01061524.1
|
|
| chr16_-_12173554 | 2.84 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr15_+_45640906 | 2.73 |
ENSDART00000149361
ENSDART00000149079 |
sagb
|
S-antigen; retina and pineal gland (arrestin) b |
| chr18_-_42830563 | 2.57 |
ENSDART00000191488
|
ttc36
|
tetratricopeptide repeat domain 36 |
| chr6_+_41096058 | 2.52 |
ENSDART00000028373
|
fkbp5
|
FK506 binding protein 5 |
| chr16_-_43971258 | 2.37 |
ENSDART00000141941
|
zfpm2a
|
zinc finger protein, FOG family member 2a |
| chr14_+_32839535 | 2.15 |
ENSDART00000168975
|
arr3b
|
arrestin 3b, retinal (X-arrestin) |
| chr9_-_9980704 | 2.06 |
ENSDART00000130243
ENSDART00000193475 |
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr7_-_24699985 | 2.01 |
ENSDART00000052802
|
calb2b
|
calbindin 2b |
| chr6_+_4539953 | 1.98 |
ENSDART00000025031
|
pou4f1
|
POU class 4 homeobox 1 |
| chr23_+_36771593 | 1.89 |
ENSDART00000078240
|
march9
|
membrane-associated ring finger (C3HC4) 9 |
| chr6_-_16406210 | 1.88 |
ENSDART00000012023
|
faimb
|
Fas apoptotic inhibitory molecule b |
| chr17_+_15882533 | 1.87 |
ENSDART00000164124
|
gabrr2a
|
gamma-aminobutyric acid (GABA) A receptor, rho 2a |
| chr22_+_20195280 | 1.83 |
ENSDART00000088603
ENSDART00000135692 |
si:dkey-110c1.7
|
si:dkey-110c1.7 |
| chr23_+_44307996 | 1.79 |
ENSDART00000042430
|
dlg4b
|
discs, large homolog 4b (Drosophila) |
| chr6_-_13187168 | 1.76 |
ENSDART00000193286
ENSDART00000188350 ENSDART00000150036 ENSDART00000149940 |
adam23a
|
ADAM metallopeptidase domain 23a |
| chr9_-_31915423 | 1.74 |
ENSDART00000060051
|
fgf14
|
fibroblast growth factor 14 |
| chr16_-_12173399 | 1.71 |
ENSDART00000142574
|
clstn3
|
calsyntenin 3 |
| chr21_-_27185915 | 1.70 |
ENSDART00000135052
|
slc8a4a
|
solute carrier family 8 (sodium/calcium exchanger), member 4a |
| chr20_-_25542183 | 1.70 |
ENSDART00000024350
|
cyp2ad2
|
cytochrome P450, family 2, subfamily AD, polypeptide 2 |
| chr13_+_33117528 | 1.65 |
ENSDART00000085719
|
si:ch211-10a23.2
|
si:ch211-10a23.2 |
| chr19_-_28367413 | 1.64 |
ENSDART00000079092
|
si:dkey-261i16.5
|
si:dkey-261i16.5 |
| chr11_-_44876005 | 1.60 |
ENSDART00000192006
|
opn6a
|
opsin 6, group member a |
| chr9_-_18877597 | 1.57 |
ENSDART00000099446
|
kctd4
|
potassium channel tetramerization domain containing 4 |
| chr1_+_1599979 | 1.50 |
ENSDART00000097626
|
urp2
|
urotensin II-related peptide |
| chr22_+_18389271 | 1.50 |
ENSDART00000088270
|
yjefn3
|
YjeF N-terminal domain containing 3 |
| chr12_+_48480632 | 1.48 |
ENSDART00000158157
|
arhgap44
|
Rho GTPase activating protein 44 |
| chr4_-_8903240 | 1.48 |
ENSDART00000129983
|
mpped1
|
metallophosphoesterase domain containing 1 |
| chr5_-_16996482 | 1.43 |
ENSDART00000144501
|
galnt9
|
polypeptide N-acetylgalactosaminyltransferase 9 |
| chr2_-_34555945 | 1.43 |
ENSDART00000056671
|
brinp2
|
bone morphogenetic protein/retinoic acid inducible neural-specific 2 |
| chr5_-_42272517 | 1.41 |
ENSDART00000137692
ENSDART00000164363 |
si:ch211-207c6.2
|
si:ch211-207c6.2 |
| chr5_-_24238733 | 1.38 |
ENSDART00000138170
|
plscr3a
|
phospholipid scramblase 3a |
| chr25_-_16057539 | 1.38 |
ENSDART00000090388
|
cyb5r2
|
cytochrome b5 reductase 2 |
| chr7_-_38633867 | 1.37 |
ENSDART00000137424
|
c1qtnf4
|
C1q and TNF related 4 |
| chr14_+_36889893 | 1.36 |
ENSDART00000124159
|
si:ch211-132p1.3
|
si:ch211-132p1.3 |
| chr25_-_13188678 | 1.34 |
ENSDART00000125754
|
si:ch211-147m6.1
|
si:ch211-147m6.1 |
| chr4_-_9592402 | 1.31 |
ENSDART00000114060
|
cdnf
|
cerebral dopamine neurotrophic factor |
| chr18_+_660578 | 1.31 |
ENSDART00000161203
|
CELF6
|
si:dkey-205h23.2 |
| chr21_+_41743493 | 1.28 |
ENSDART00000192669
|
ppp2r2bb
|
protein phosphatase 2, regulatory subunit B, beta b |
| chr2_+_24203229 | 1.28 |
ENSDART00000138088
|
map4l
|
microtubule associated protein 4 like |
| chr22_+_5106751 | 1.27 |
ENSDART00000138967
|
atcaya
|
ataxia, cerebellar, Cayman type a |
| chr3_-_32337653 | 1.26 |
ENSDART00000156918
ENSDART00000156551 |
si:dkey-16p21.8
|
si:dkey-16p21.8 |
| chr6_-_13188667 | 1.26 |
ENSDART00000191654
|
adam23a
|
ADAM metallopeptidase domain 23a |
| chr11_+_25634041 | 1.25 |
ENSDART00000033657
|
grm6b
|
glutamate receptor, metabotropic 6b |
| chr6_+_23935045 | 1.25 |
ENSDART00000162993
|
gadd45ab
|
growth arrest and DNA-damage-inducible, alpha, b |
| chr19_-_32042105 | 1.24 |
ENSDART00000088358
|
znf704
|
zinc finger protein 704 |
| chr5_-_24000211 | 1.22 |
ENSDART00000188865
|
map7d2a
|
MAP7 domain containing 2a |
| chr20_+_18225329 | 1.21 |
ENSDART00000144172
|
kctd1
|
potassium channel tetramerization domain containing 1 |
| chr23_+_28731379 | 1.18 |
ENSDART00000047378
|
cort
|
cortistatin |
| chr5_-_25406807 | 1.18 |
ENSDART00000089748
|
rorb
|
RAR-related orphan receptor B |
| chr16_+_45746549 | 1.17 |
ENSDART00000190403
|
paqr6
|
progestin and adipoQ receptor family member VI |
| chr10_+_26972755 | 1.15 |
ENSDART00000042162
|
tm7sf2
|
transmembrane 7 superfamily member 2 |
| chr22_+_14051894 | 1.15 |
ENSDART00000142548
|
aox6
|
aldehyde oxidase 6 |
| chr6_+_9867426 | 1.13 |
ENSDART00000151749
|
MPP4 (1 of many)
|
si:ch211-222n4.6 |
| chr24_-_24983047 | 1.13 |
ENSDART00000066631
|
slc51a
|
solute carrier family 51, alpha subunit |
| chr10_+_453619 | 1.11 |
ENSDART00000135598
|
zdhhc8a
|
zinc finger, DHHC-type containing 8a |
| chr5_+_27429872 | 1.11 |
ENSDART00000087894
|
zgc:165555
|
zgc:165555 |
| chr15_-_30815826 | 1.11 |
ENSDART00000156160
ENSDART00000145918 |
msi2b
|
musashi RNA-binding protein 2b |
| chr25_+_27923846 | 1.10 |
ENSDART00000047007
|
slc13a1
|
solute carrier family 13 member 1 |
| chr2_+_55982940 | 1.10 |
ENSDART00000097753
ENSDART00000097751 |
nmrk2
|
nicotinamide riboside kinase 2 |
| chr12_+_46960579 | 1.10 |
ENSDART00000149032
|
oat
|
ornithine aminotransferase |
| chr10_-_35103208 | 1.08 |
ENSDART00000192734
|
zgc:110006
|
zgc:110006 |
| chr3_-_32817274 | 1.07 |
ENSDART00000142582
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr20_+_40150612 | 1.06 |
ENSDART00000143680
ENSDART00000109681 ENSDART00000101041 ENSDART00000121818 |
trdn
|
triadin |
| chr22_-_18491813 | 1.05 |
ENSDART00000105419
|
si:ch211-212d10.2
|
si:ch211-212d10.2 |
| chr23_+_28582865 | 1.04 |
ENSDART00000020296
|
l1cama
|
L1 cell adhesion molecule, paralog a |
| chr2_+_30147504 | 1.04 |
ENSDART00000190947
|
kcnb2
|
potassium voltage-gated channel, Shab-related subfamily, member 2 |
| chr18_-_36135799 | 1.04 |
ENSDART00000059344
|
b3gat1a
|
beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P) a |
| chr7_+_13824150 | 1.02 |
ENSDART00000035067
|
abhd2a
|
abhydrolase domain containing 2a |
| chr24_-_40009446 | 0.99 |
ENSDART00000087422
|
aoc1
|
amine oxidase, copper containing 1 |
| chr4_+_12292274 | 0.96 |
ENSDART00000061070
ENSDART00000150786 |
mkrn1
|
makorin, ring finger protein, 1 |
| chr18_+_7286788 | 0.95 |
ENSDART00000022998
|
ANO2 (1 of many)
|
si:ch73-86n2.1 |
| chr22_-_7400471 | 0.95 |
ENSDART00000161215
|
si:dkey-57c15.1
|
si:dkey-57c15.1 |
| chr2_+_32835679 | 0.91 |
ENSDART00000132767
|
pxdc1a
|
PX domain containing 1a |
| chr10_+_22003750 | 0.91 |
ENSDART00000109420
|
kcnip1b
|
Kv channel interacting protein 1 b |
| chr25_-_25736958 | 0.91 |
ENSDART00000166308
|
cib2
|
calcium and integrin binding family member 2 |
| chr8_+_13503377 | 0.91 |
ENSDART00000034740
ENSDART00000167187 |
fut9d
|
fucosyltransferase 9d |
| chr21_-_13662237 | 0.90 |
ENSDART00000091647
ENSDART00000151547 |
pnpla7a
|
patatin-like phospholipase domain containing 7a |
| chr10_-_24689725 | 0.89 |
ENSDART00000079566
|
si:ch211-287a12.9
|
si:ch211-287a12.9 |
| chr13_+_30951155 | 0.86 |
ENSDART00000057469
ENSDART00000162254 |
vstm4a
|
V-set and transmembrane domain containing 4a |
| chr12_-_28983584 | 0.86 |
ENSDART00000112374
|
zgc:171713
|
zgc:171713 |
| chr8_-_15398760 | 0.83 |
ENSDART00000162011
|
agbl4
|
ATP/GTP binding protein-like 4 |
| chr4_-_16644708 | 0.82 |
ENSDART00000042307
|
sinhcaf
|
SIN3-HDAC complex associated factor |
| chr19_+_3001473 | 0.78 |
ENSDART00000145363
|
scxa
|
scleraxis bHLH transcription factor a |
| chr5_+_30384554 | 0.77 |
ENSDART00000135483
|
zgc:158412
|
zgc:158412 |
| chr6_+_45692026 | 0.76 |
ENSDART00000164759
|
cntn4
|
contactin 4 |
| chr10_+_33744098 | 0.75 |
ENSDART00000147775
|
rxfp2a
|
relaxin/insulin-like family peptide receptor 2a |
| chr25_-_19666107 | 0.72 |
ENSDART00000149889
|
atp2b1b
|
ATPase plasma membrane Ca2+ transporting 1b |
| chr19_+_14573998 | 0.72 |
ENSDART00000022076
|
fam46bb
|
family with sequence similarity 46, member Bb |
| chr4_+_12013043 | 0.71 |
ENSDART00000130692
|
cry1aa
|
cryptochrome circadian clock 1aa |
| chr8_+_31777633 | 0.71 |
ENSDART00000142519
|
oxct1a
|
3-oxoacid CoA transferase 1a |
| chr3_-_3209432 | 0.70 |
ENSDART00000140635
|
si:ch211-229i14.2
|
si:ch211-229i14.2 |
| chr15_-_5157572 | 0.69 |
ENSDART00000174192
|
or128-10
|
odorant receptor, family E, subfamily 128, member 10 |
| chr4_+_12013642 | 0.68 |
ENSDART00000067281
|
cry1aa
|
cryptochrome circadian clock 1aa |
| chr2_+_2563192 | 0.68 |
ENSDART00000055713
|
CABZ01033000.1
|
|
| chr19_-_8096984 | 0.67 |
ENSDART00000146987
|
si:dkey-266f7.9
|
si:dkey-266f7.9 |
| chr20_+_35998073 | 0.67 |
ENSDART00000140457
|
opn5
|
opsin 5 |
| chr25_-_26673570 | 0.67 |
ENSDART00000154917
|
ciartb
|
circadian associated repressor of transcription b |
| chr17_-_30473374 | 0.67 |
ENSDART00000155021
|
si:ch211-175f11.5
|
si:ch211-175f11.5 |
| chr22_+_24645325 | 0.67 |
ENSDART00000159531
|
lpar3
|
lysophosphatidic acid receptor 3 |
| chr13_-_49444636 | 0.65 |
ENSDART00000136991
|
irf2bp2a
|
interferon regulatory factor 2 binding protein 2a |
| chr20_+_27298783 | 0.63 |
ENSDART00000013861
|
ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chr10_+_44940693 | 0.63 |
ENSDART00000157515
|
cnnm4a
|
cyclin and CBS domain divalent metal cation transport mediator 4a |
| chr19_-_30562648 | 0.63 |
ENSDART00000171006
|
hpcal4
|
hippocalcin like 4 |
| chr15_+_39096736 | 0.62 |
ENSDART00000129511
ENSDART00000014877 |
robo2
|
roundabout, axon guidance receptor, homolog 2 (Drosophila) |
| chr22_-_38516922 | 0.59 |
ENSDART00000151257
ENSDART00000151785 |
klc4
|
kinesin light chain 4 |
| chr17_+_33719415 | 0.59 |
ENSDART00000132294
|
fut8a
|
fucosyltransferase 8a (alpha (1,6) fucosyltransferase) |
| chr15_+_42933236 | 0.58 |
ENSDART00000167763
|
slc8a2b
|
solute carrier family 8 (sodium/calcium exchanger), member 2b |
| chr25_+_20188769 | 0.58 |
ENSDART00000142781
|
cald1b
|
caldesmon 1b |
| chr7_-_6466119 | 0.57 |
ENSDART00000173138
|
zgc:112234
|
zgc:112234 |
| chr22_+_18786797 | 0.56 |
ENSDART00000141864
|
cbarpb
|
calcium channel, voltage-dependent, beta subunit associated regulatory protein b |
| chr20_-_39188476 | 0.56 |
ENSDART00000152858
|
rcan2
|
regulator of calcineurin 2 |
| chr22_+_17784414 | 0.56 |
ENSDART00000188189
ENSDART00000145260 |
tm6sf2
|
transmembrane 6 superfamily member 2 |
| chr9_-_18735256 | 0.55 |
ENSDART00000143165
|
tsc22d1
|
TSC22 domain family, member 1 |
| chr14_+_23874062 | 0.55 |
ENSDART00000172149
|
sh3rf2
|
SH3 domain containing ring finger 2 |
| chr16_+_39159752 | 0.54 |
ENSDART00000122081
|
sybu
|
syntabulin (syntaxin-interacting) |
| chr7_+_69470442 | 0.54 |
ENSDART00000189593
|
gabarapb
|
GABA(A) receptor-associated protein b |
| chr12_+_36092218 | 0.54 |
ENSDART00000168694
|
btbd17a
|
BTB (POZ) domain containing 17a |
| chr25_+_35067318 | 0.54 |
ENSDART00000186828
|
CU302436.1
|
|
| chr21_+_8427059 | 0.54 |
ENSDART00000143151
|
dennd1a
|
DENN/MADD domain containing 1A |
| chr7_+_61050329 | 0.54 |
ENSDART00000115355
|
nwd2
|
NACHT and WD repeat domain containing 2 |
| chr11_+_769623 | 0.54 |
ENSDART00000172972
|
vgll4b
|
vestigial-like family member 4b |
| chr5_+_30624183 | 0.53 |
ENSDART00000141444
|
abcg4a
|
ATP-binding cassette, sub-family G (WHITE), member 4a |
| chr7_-_4036184 | 0.53 |
ENSDART00000019949
|
ndrg2
|
NDRG family member 2 |
| chr4_+_20255160 | 0.50 |
ENSDART00000188658
|
lrtm2a
|
leucine-rich repeats and transmembrane domains 2a |
| chr10_+_26973063 | 0.50 |
ENSDART00000143162
ENSDART00000186210 |
tm7sf2
|
transmembrane 7 superfamily member 2 |
| chr7_+_66739166 | 0.49 |
ENSDART00000154634
|
mrvi1
|
murine retrovirus integration site 1 homolog |
| chr7_+_5906327 | 0.49 |
ENSDART00000173160
|
zgc:112234
|
zgc:112234 |
| chr7_+_27455321 | 0.49 |
ENSDART00000148417
|
cyp2r1
|
cytochrome P450, family 2, subfamily R, polypeptide 1 |
| chr5_-_46896541 | 0.49 |
ENSDART00000133240
|
edil3a
|
EGF-like repeats and discoidin I-like domains 3a |
| chr10_+_36458563 | 0.48 |
ENSDART00000077008
|
alox5ap
|
arachidonate 5-lipoxygenase-activating protein |
| chr10_-_5821584 | 0.48 |
ENSDART00000166388
|
il6st
|
interleukin 6 signal transducer |
| chr5_+_29820266 | 0.47 |
ENSDART00000146331
ENSDART00000098315 |
f11r.2
|
F11 receptor, tandem duplicate 2 |
| chr5_+_5398966 | 0.47 |
ENSDART00000139553
|
mapkap1
|
mitogen-activated protein kinase associated protein 1 |
| chr7_-_6351021 | 0.47 |
ENSDART00000159542
|
zgc:112234
|
zgc:112234 |
| chr23_-_893547 | 0.47 |
ENSDART00000136805
|
rbm10
|
RNA binding motif protein 10 |
| chr21_-_35853245 | 0.47 |
ENSDART00000172245
|
sgcd
|
sarcoglycan, delta (dystrophin-associated glycoprotein) |
| chr11_-_41130239 | 0.46 |
ENSDART00000173268
|
dnajc11b
|
DnaJ (Hsp40) homolog, subfamily C, member 11b |
| chr18_-_49286381 | 0.45 |
ENSDART00000174248
ENSDART00000174038 |
si:zfos-464b6.2
|
si:zfos-464b6.2 |
| chr2_-_39036604 | 0.45 |
ENSDART00000129963
|
rbp1
|
retinol binding protein 1b, cellular |
| chr4_-_15603511 | 0.44 |
ENSDART00000122520
ENSDART00000162356 |
chchd3a
|
coiled-coil-helix-coiled-coil-helix domain containing 3a |
| chr2_-_3616363 | 0.42 |
ENSDART00000144699
|
pter
|
phosphotriesterase related |
| chr16_+_40560622 | 0.42 |
ENSDART00000038294
|
tp53inp1
|
tumor protein p53 inducible nuclear protein 1 |
| chr19_-_19556778 | 0.41 |
ENSDART00000164060
|
tax1bp1a
|
Tax1 (human T-cell leukemia virus type I) binding protein 1a |
| chr3_+_47322494 | 0.41 |
ENSDART00000102202
|
ppap2d
|
phosphatidic acid phosphatase type 2D |
| chr20_-_39735952 | 0.40 |
ENSDART00000101049
ENSDART00000137485 ENSDART00000062402 |
tpd52l1
|
tumor protein D52-like 1 |
| chr14_-_16810401 | 0.40 |
ENSDART00000158396
ENSDART00000170758 |
tcirg1b
|
T cell immune regulator 1, ATPase H+ transporting V0 subunit a3b |
| chr9_+_45227028 | 0.40 |
ENSDART00000185579
|
adarb1b
|
adenosine deaminase, RNA-specific, B1b |
| chr8_-_12403077 | 0.39 |
ENSDART00000142150
|
phf19
|
PHD finger protein 19 |
| chr18_-_47103131 | 0.39 |
ENSDART00000188553
|
CABZ01039304.1
|
|
| chr13_+_42406883 | 0.39 |
ENSDART00000084354
|
cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr25_+_36674715 | 0.38 |
ENSDART00000111861
|
mafb
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog b (paralog b) |
| chr10_+_33393829 | 0.37 |
ENSDART00000163458
ENSDART00000115379 |
zgc:153345
|
zgc:153345 |
| chr15_+_34592215 | 0.37 |
ENSDART00000099776
|
tspan13a
|
tetraspanin 13a |
| chr20_-_45060241 | 0.37 |
ENSDART00000185227
|
klhl29
|
kelch-like family member 29 |
| chr21_-_30254185 | 0.37 |
ENSDART00000101054
|
dnajc18
|
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr22_-_20376488 | 0.36 |
ENSDART00000140187
|
zbtb7a
|
zinc finger and BTB domain containing 7a |
| chr6_-_52644182 | 0.35 |
ENSDART00000015051
|
gdf5
|
growth differentiation factor 5 |
| chr21_-_25573064 | 0.33 |
ENSDART00000134310
|
CR388166.1
|
|
| chr15_-_734203 | 0.33 |
ENSDART00000177867
ENSDART00000154918 |
si:dkey-7i4.21
BX004964.1
|
si:dkey-7i4.21 |
| chr6_-_13308813 | 0.33 |
ENSDART00000065372
|
kcnj3b
|
potassium inwardly-rectifying channel, subfamily J, member 3b |
| chr7_+_10701938 | 0.33 |
ENSDART00000158162
|
arnt2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr17_-_15528597 | 0.33 |
ENSDART00000150232
|
fyna
|
FYN proto-oncogene, Src family tyrosine kinase a |
| chr3_+_26223376 | 0.32 |
ENSDART00000128284
|
nudt9
|
nudix (nucleoside diphosphate linked moiety X)-type motif 9 |
| chr17_-_32865788 | 0.31 |
ENSDART00000077476
|
prox1a
|
prospero homeobox 1a |
| chr18_-_35390735 | 0.31 |
ENSDART00000098297
|
CABZ01040999.1
|
|
| chr3_+_33345348 | 0.30 |
ENSDART00000059262
|
mpp2a
|
membrane protein, palmitoylated 2a (MAGUK p55 subfamily member 2) |
| chr25_+_5604512 | 0.30 |
ENSDART00000042781
|
plxnb2b
|
plexin b2b |
| chr8_+_18044257 | 0.30 |
ENSDART00000029255
|
glis1b
|
GLIS family zinc finger 1b |
| chr24_+_41931585 | 0.29 |
ENSDART00000130310
|
epb41l3a
|
erythrocyte membrane protein band 4.1-like 3a |
| chr15_-_4023182 | 0.29 |
ENSDART00000190195
|
CR626884.5
|
|
| chr17_-_24916889 | 0.29 |
ENSDART00000156157
|
si:ch211-195o20.7
|
si:ch211-195o20.7 |
| chr1_-_16394814 | 0.29 |
ENSDART00000013024
|
fgf20a
|
fibroblast growth factor 20a |
| chr14_+_6991142 | 0.28 |
ENSDART00000157635
ENSDART00000059918 |
hnrnph1
|
heterogeneous nuclear ribonucleoprotein H1 |
| chr16_-_31756859 | 0.28 |
ENSDART00000149170
ENSDART00000126617 ENSDART00000182722 |
ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr1_-_55068941 | 0.28 |
ENSDART00000152143
ENSDART00000152590 |
peli1a
|
pellino E3 ubiquitin protein ligase 1a |
| chr7_+_31074636 | 0.28 |
ENSDART00000173805
|
tjp1a
|
tight junction protein 1a |
| chr25_-_13202208 | 0.28 |
ENSDART00000168155
|
si:ch211-194m7.4
|
si:ch211-194m7.4 |
| chr2_+_8779164 | 0.28 |
ENSDART00000134308
|
zzz3
|
zinc finger, ZZ-type containing 3 |
| chr9_-_25444091 | 0.27 |
ENSDART00000002428
|
acvr2aa
|
activin A receptor type 2Aa |
| chr1_+_8984068 | 0.27 |
ENSDART00000122568
|
tlr2
|
toll-like receptor 2 |
| chr19_-_6239248 | 0.27 |
ENSDART00000014127
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr23_-_6765653 | 0.26 |
ENSDART00000192310
|
FP102169.1
|
|
| chr25_+_14694876 | 0.26 |
ENSDART00000050478
ENSDART00000188093 |
mpped2
|
metallophosphoesterase domain containing 2b |
| chr9_+_54695981 | 0.25 |
ENSDART00000183605
|
rab9a
|
RAB9A, member RAS oncogene family |
| chr7_+_44593756 | 0.25 |
ENSDART00000125365
|
si:ch211-189a15.5
|
si:ch211-189a15.5 |
| chr3_+_16922226 | 0.24 |
ENSDART00000017646
|
atp6v0a1a
|
ATPase H+ transporting V0 subunit a1a |
| chr7_-_33921366 | 0.24 |
ENSDART00000052397
|
pias1a
|
protein inhibitor of activated STAT, 1a |
| chr22_-_32392252 | 0.23 |
ENSDART00000148681
ENSDART00000137945 |
pcbp4
|
poly(rC) binding protein 4 |
| chr19_+_30990129 | 0.22 |
ENSDART00000052169
ENSDART00000193376 |
sync
|
syncoilin, intermediate filament protein |
| chr5_+_22970617 | 0.22 |
ENSDART00000192859
|
hmgn7
|
high mobility group nucleosomal binding domain 7 |
| chr2_-_56131312 | 0.21 |
ENSDART00000097755
|
jund
|
JunD proto-oncogene, AP-1 transcription factor subunit |
| chr10_-_28477023 | 0.21 |
ENSDART00000137008
|
bbx
|
bobby sox homolog (Drosophila) |
| chr3_+_2669813 | 0.21 |
ENSDART00000014205
|
CR388047.1
|
|
| chr14_-_28001986 | 0.20 |
ENSDART00000054115
|
tsc22d3
|
TSC22 domain family, member 3 |
| chr19_+_30990815 | 0.20 |
ENSDART00000134645
|
sync
|
syncoilin, intermediate filament protein |
| chr16_+_43152727 | 0.20 |
ENSDART00000125590
ENSDART00000154493 |
adam22
|
ADAM metallopeptidase domain 22 |
| chr11_-_45216684 | 0.19 |
ENSDART00000168621
|
tmc8
|
transmembrane channel-like 8 |
| chr19_+_41464870 | 0.19 |
ENSDART00000102778
|
dlx6a
|
distal-less homeobox 6a |
Network of associatons between targets according to the STRING database.
First level regulatory network of pou3f1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0097688 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.5 | 4.5 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.5 | 2.0 | GO:0099509 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.4 | 1.5 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.3 | 4.1 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.3 | 1.8 | GO:0099525 | synaptic vesicle docking(GO:0016081) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.3 | 0.8 | GO:0035992 | tendon formation(GO:0035992) |
| 0.2 | 0.9 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.2 | 0.7 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.2 | 1.0 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.2 | 1.0 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.2 | 1.4 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.2 | 1.9 | GO:1902041 | regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902041) negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.2 | 0.7 | GO:0016038 | absorption of visible light(GO:0016038) |
| 0.2 | 0.5 | GO:0030952 | establishment or maintenance of cytoskeleton polarity(GO:0030952) |
| 0.1 | 0.3 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.1 | 1.7 | GO:0042044 | fluid transport(GO:0042044) |
| 0.1 | 0.7 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 1.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.5 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.1 | 1.3 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 1.7 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 1.1 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 1.1 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.5 | GO:0070572 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.1 | 1.3 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.1 | 0.7 | GO:0071305 | vitamin D3 metabolic process(GO:0070640) cellular response to vitamin D(GO:0071305) |
| 0.1 | 1.0 | GO:0071941 | nitrogen cycle metabolic process(GO:0071941) |
| 0.1 | 1.2 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 2.9 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.1 | 0.5 | GO:0019370 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.1 | 1.5 | GO:0036065 | fucosylation(GO:0036065) |
| 0.1 | 0.3 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) |
| 0.1 | 2.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 1.1 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 0.6 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) negative regulation of regulated secretory pathway(GO:1903306) |
| 0.1 | 0.6 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 3.0 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.1 | 1.6 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.1 | 1.0 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.1 | 0.6 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.1 | 1.3 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 1.0 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 0.8 | GO:0030819 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.1 | 0.3 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.1 | 0.6 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.6 | GO:0045851 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.0 | 0.2 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.5 | GO:0048796 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.0 | 1.4 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 1.1 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.3 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 1.0 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 0.1 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.5 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.0 | 1.7 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.5 | GO:0090559 | intestinal absorption(GO:0050892) regulation of membrane permeability(GO:0090559) |
| 0.0 | 0.4 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.6 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 1.6 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.9 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) |
| 0.0 | 0.5 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.0 | 0.4 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.8 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.3 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.0 | 0.5 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.2 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 1.9 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.2 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 2.8 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.4 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.3 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.0 | 0.4 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.0 | 1.5 | GO:0008217 | regulation of blood pressure(GO:0008217) |
| 0.0 | 0.5 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:0050654 | chondroitin sulfate proteoglycan metabolic process(GO:0050654) |
| 0.0 | 0.4 | GO:0071218 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.0 | 0.7 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.1 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.0 | 0.1 | GO:0051932 | regulation of synaptic transmission, GABAergic(GO:0032228) synaptic transmission, GABAergic(GO:0051932) |
| 0.0 | 0.1 | GO:0060231 | mesenchymal to epithelial transition(GO:0060231) |
| 0.0 | 0.6 | GO:0043534 | blood vessel endothelial cell migration(GO:0043534) |
| 0.0 | 0.2 | GO:0035141 | medial fin morphogenesis(GO:0035141) |
| 0.0 | 0.1 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 1.1 | GO:0006352 | DNA-templated transcription, initiation(GO:0006352) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.1 | GO:0098894 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.3 | 1.0 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.2 | 3.8 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 2.7 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 1.0 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.9 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 1.8 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 0.8 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 0.5 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.6 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 1.9 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 1.1 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 1.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.7 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 1.1 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.3 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.3 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 1.3 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 2.3 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 1.5 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.5 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 4.3 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.3 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.5 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 1.4 | GO:0043025 | neuronal cell body(GO:0043025) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.5 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.4 | 1.2 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.3 | 1.0 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.3 | 1.3 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.2 | 3.1 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.2 | 0.7 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.2 | 1.0 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 1.0 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.2 | 1.4 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.2 | 1.2 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.2 | 2.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.2 | 1.0 | GO:0008126 | acetylesterase activity(GO:0008126) |
| 0.2 | 1.1 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.1 | 1.0 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.1 | 0.7 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.1 | 0.5 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.1 | 1.3 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.1 | 1.4 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.5 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 0.6 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 1.5 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.1 | 0.9 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 1.1 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 1.5 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.1 | 1.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 1.7 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 2.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 1.9 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 0.6 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 1.1 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.7 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.6 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 1.2 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 2.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 2.4 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.4 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 1.6 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.7 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.5 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.8 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 2.2 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.4 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.0 | 0.7 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.4 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.3 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.2 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 1.9 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 2.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 3.1 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 0.3 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.4 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 2.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 1.6 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.4 | GO:0035064 | methylated histone binding(GO:0035064) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 2.5 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.5 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.5 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.8 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 1.1 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 0.3 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.1 | 0.3 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 0.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 1.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.7 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.5 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |