Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
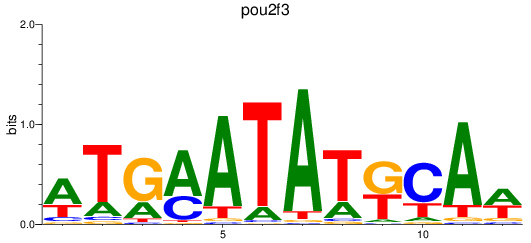
Results for pou2f3
Z-value: 0.97
Transcription factors associated with pou2f3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pou2f3
|
ENSDARG00000052387 | POU class 2 homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pou2f3 | dr11_v1_chr5_+_58550795_58550795 | 0.44 | 6.0e-02 | Click! |
Activity profile of pou2f3 motif
Sorted Z-values of pou2f3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_14052349 | 3.99 |
ENSDART00000135811
|
atp2b3a
|
ATPase plasma membrane Ca2+ transporting 3a |
| chr17_-_37395460 | 3.10 |
ENSDART00000148160
ENSDART00000075975 |
crip1
|
cysteine-rich protein 1 |
| chr16_+_26012569 | 3.05 |
ENSDART00000148846
|
prss59.1
|
protease, serine, 59, tandem duplicate 1 |
| chr25_+_35189555 | 3.03 |
ENSDART00000044453
|
ano5a
|
anoctamin 5a |
| chr25_+_31277415 | 2.94 |
ENSDART00000036275
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr9_+_49659114 | 2.90 |
ENSDART00000189643
|
CSRNP3
|
cysteine and serine rich nuclear protein 3 |
| chr14_+_15155684 | 2.88 |
ENSDART00000167966
|
zgc:158852
|
zgc:158852 |
| chr5_-_6567464 | 2.82 |
ENSDART00000184985
|
tnks1bp1
|
tankyrase 1 binding protein 1 |
| chr14_-_46238186 | 2.80 |
ENSDART00000173245
|
si:ch211-113d11.6
|
si:ch211-113d11.6 |
| chr16_+_2820340 | 2.71 |
ENSDART00000092299
ENSDART00000192931 ENSDART00000148512 |
si:dkey-288i20.2
|
si:dkey-288i20.2 |
| chr18_-_20869175 | 2.66 |
ENSDART00000090079
|
synm
|
synemin, intermediate filament protein |
| chr9_-_22158325 | 2.48 |
ENSDART00000114568
|
crygm2d16
|
crystallin, gamma M2d16 |
| chr9_-_22129788 | 2.46 |
ENSDART00000124272
ENSDART00000175417 |
crygm2d8
|
crystallin, gamma M2d8 |
| chr8_+_22931427 | 2.43 |
ENSDART00000063096
|
sypa
|
synaptophysin a |
| chr24_+_402493 | 2.32 |
ENSDART00000036472
|
VSTM2A
|
zgc:110852 |
| chr6_+_6924637 | 2.29 |
ENSDART00000065551
ENSDART00000151393 |
zak
|
sterile alpha motif and leucine zipper containing kinase AZK |
| chr25_+_31227747 | 2.24 |
ENSDART00000033872
|
tnni2a.1
|
troponin I type 2a (skeletal, fast), tandem duplicate 1 |
| chr4_-_8903240 | 2.23 |
ENSDART00000129983
|
mpped1
|
metallophosphoesterase domain containing 1 |
| chr16_-_52540056 | 2.15 |
ENSDART00000188304
|
CR293507.1
|
|
| chr21_-_37733571 | 2.15 |
ENSDART00000176214
|
mpp1
|
membrane protein, palmitoylated 1 |
| chr11_-_762721 | 2.03 |
ENSDART00000166465
|
syn2b
|
synapsin IIb |
| chr19_-_205104 | 2.03 |
ENSDART00000011890
|
zbtb22a
|
zinc finger and BTB domain containing 22a |
| chr12_-_21834611 | 2.00 |
ENSDART00000179553
|
thrab
|
thyroid hormone receptor alpha b |
| chr16_+_46111849 | 1.98 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr11_+_37275448 | 1.95 |
ENSDART00000161423
|
creld1a
|
cysteine-rich with EGF-like domains 1a |
| chr13_+_24022963 | 1.88 |
ENSDART00000028285
|
pgbd5
|
piggyBac transposable element derived 5 |
| chr13_-_2189761 | 1.88 |
ENSDART00000166255
|
mlip
|
muscular LMNA-interacting protein |
| chr3_+_45687266 | 1.83 |
ENSDART00000131652
|
gpr146
|
G protein-coupled receptor 146 |
| chr9_-_9982696 | 1.80 |
ENSDART00000192548
ENSDART00000125852 |
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr24_-_7321928 | 1.80 |
ENSDART00000167570
ENSDART00000045150 |
actr3b
|
ARP3 actin related protein 3 homolog B |
| chr18_+_3579829 | 1.80 |
ENSDART00000158763
ENSDART00000182850 ENSDART00000162754 ENSDART00000178789 ENSDART00000172656 |
lrch3
|
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr22_+_18389271 | 1.75 |
ENSDART00000088270
|
yjefn3
|
YjeF N-terminal domain containing 3 |
| chr1_+_25801648 | 1.71 |
ENSDART00000129471
|
gucy1b1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr6_-_54796072 | 1.69 |
ENSDART00000164445
|
tnnt2b
|
troponin T type 2b (cardiac) |
| chr25_+_24291156 | 1.67 |
ENSDART00000083407
|
b4galnt4a
|
beta-1,4-N-acetyl-galactosaminyl transferase 4a |
| chr5_+_4332220 | 1.67 |
ENSDART00000051699
|
sat1a.1
|
spermidine/spermine N1-acetyltransferase 1a, duplicate 1 |
| chr13_+_2442841 | 1.66 |
ENSDART00000114456
ENSDART00000137124 ENSDART00000193737 ENSDART00000189722 ENSDART00000187485 |
arfgef3
|
ARFGEF family member 3 |
| chr25_+_37285737 | 1.64 |
ENSDART00000126879
|
tmed6
|
transmembrane p24 trafficking protein 6 |
| chr19_-_5345930 | 1.62 |
ENSDART00000066620
ENSDART00000151398 |
krtt1c19e
|
keratin type 1 c19e |
| chr3_-_46410387 | 1.57 |
ENSDART00000156822
|
cdip1
|
cell death-inducing p53 target 1 |
| chr19_+_1510971 | 1.56 |
ENSDART00000157721
|
SLC45A4 (1 of many)
|
solute carrier family 45 member 4 |
| chr23_-_35066816 | 1.55 |
ENSDART00000168731
ENSDART00000163731 |
BX294434.1
|
|
| chr16_-_12173554 | 1.54 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr19_+_5076617 | 1.49 |
ENSDART00000189990
|
eno2
|
enolase 2 |
| chr20_-_4031475 | 1.47 |
ENSDART00000112053
|
fam89a
|
family with sequence similarity 89, member A |
| chr22_+_14051894 | 1.41 |
ENSDART00000142548
|
aox6
|
aldehyde oxidase 6 |
| chr20_-_46817223 | 1.40 |
ENSDART00000100336
|
esrrgb
|
estrogen-related receptor gamma b |
| chr2_-_52455525 | 1.40 |
ENSDART00000160768
ENSDART00000002241 |
chico
|
chico |
| chr2_-_1443692 | 1.38 |
ENSDART00000004533
|
rpe65a
|
retinal pigment epithelium-specific protein 65a |
| chr19_-_5103313 | 1.38 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr14_+_32839535 | 1.37 |
ENSDART00000168975
|
arr3b
|
arrestin 3b, retinal (X-arrestin) |
| chr3_+_22327738 | 1.36 |
ENSDART00000055675
|
gh1
|
growth hormone 1 |
| chr11_+_30817943 | 1.36 |
ENSDART00000150130
ENSDART00000159997 |
cacna1ab
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit, b |
| chr1_-_21901589 | 1.34 |
ENSDART00000140553
|
frmpd1a
|
FERM and PDZ domain containing 1a |
| chr12_+_48480632 | 1.33 |
ENSDART00000158157
|
arhgap44
|
Rho GTPase activating protein 44 |
| chr16_-_12173399 | 1.31 |
ENSDART00000142574
|
clstn3
|
calsyntenin 3 |
| chr6_+_49722970 | 1.30 |
ENSDART00000155934
ENSDART00000154738 |
stx16
|
syntaxin 16 |
| chr9_-_30165621 | 1.29 |
ENSDART00000089543
|
abi3bpa
|
ABI family, member 3 (NESH) binding protein a |
| chr25_-_3503458 | 1.27 |
ENSDART00000173269
|
PRKAR2B
|
si:ch211-272n13.7 |
| chr25_-_25058508 | 1.26 |
ENSDART00000087570
ENSDART00000178891 |
FQ311928.1
|
|
| chr20_+_9128829 | 1.24 |
ENSDART00000064144
ENSDART00000137450 |
bpnt1
|
bisphosphate nucleotidase 1 |
| chr11_+_30000814 | 1.24 |
ENSDART00000191011
ENSDART00000189770 |
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr20_-_39789036 | 1.24 |
ENSDART00000086405
ENSDART00000098253 |
rnf217
|
ring finger protein 217 |
| chr18_+_7283283 | 1.24 |
ENSDART00000141493
|
ANO2 (1 of many)
|
si:ch73-86n2.1 |
| chr4_+_25950372 | 1.24 |
ENSDART00000125767
|
metap2a
|
methionyl aminopeptidase 2a |
| chr20_-_14206698 | 1.23 |
ENSDART00000082441
|
dlgap2b
|
discs, large (Drosophila) homolog-associated protein 2b |
| chr16_+_14029283 | 1.22 |
ENSDART00000146165
ENSDART00000132075 |
rusc1
|
RUN and SH3 domain containing 1 |
| chr3_-_5067585 | 1.21 |
ENSDART00000169609
|
tefb
|
thyrotrophic embryonic factor b |
| chr5_-_64883082 | 1.20 |
ENSDART00000064983
ENSDART00000139066 |
krt1-c5
|
keratin, type 1, gene c5 |
| chr16_+_50089417 | 1.19 |
ENSDART00000153675
|
nr1d2a
|
nuclear receptor subfamily 1, group D, member 2a |
| chr11_-_37880492 | 1.19 |
ENSDART00000102868
|
etnk2
|
ethanolamine kinase 2 |
| chr19_-_8096984 | 1.18 |
ENSDART00000146987
|
si:dkey-266f7.9
|
si:dkey-266f7.9 |
| chr3_+_59851537 | 1.17 |
ENSDART00000180997
|
CU693479.1
|
|
| chr5_-_13206878 | 1.16 |
ENSDART00000051666
|
ppm1f
|
protein phosphatase, Mg2+/Mn2+ dependent, 1F |
| chr6_+_49723289 | 1.16 |
ENSDART00000190452
|
stx16
|
syntaxin 16 |
| chr6_+_55174744 | 1.16 |
ENSDART00000023562
|
SYT2
|
synaptotagmin 2 |
| chr20_+_41664350 | 1.15 |
ENSDART00000186247
|
fam184a
|
family with sequence similarity 184, member A |
| chr8_-_7444390 | 1.14 |
ENSDART00000149671
|
hdac6
|
histone deacetylase 6 |
| chr13_-_2215213 | 1.13 |
ENSDART00000129773
|
mlip
|
muscular LMNA-interacting protein |
| chr24_-_24849091 | 1.13 |
ENSDART00000133649
ENSDART00000038290 |
crhb
|
corticotropin releasing hormone b |
| chr14_+_597532 | 1.13 |
ENSDART00000159805
|
LO018568.2
|
|
| chr24_-_41220538 | 1.11 |
ENSDART00000150207
|
acvr2ba
|
activin A receptor type 2Ba |
| chr6_+_51773873 | 1.09 |
ENSDART00000156516
|
tmem74b
|
transmembrane protein 74B |
| chr14_-_9713549 | 1.07 |
ENSDART00000193356
ENSDART00000166739 |
si:zfos-2326c3.2
|
si:zfos-2326c3.2 |
| chr23_-_27345425 | 1.07 |
ENSDART00000022042
ENSDART00000191870 |
scn8aa
|
sodium channel, voltage gated, type VIII, alpha subunit a |
| chr24_+_792429 | 1.06 |
ENSDART00000082523
|
impa2
|
inositol(myo)-1(or 4)-monophosphatase 2 |
| chr7_-_18168493 | 1.06 |
ENSDART00000127428
|
peli3
|
pellino E3 ubiquitin protein ligase family member 3 |
| chr14_-_2213660 | 1.05 |
ENSDART00000162537
|
pcdh2ab3
|
protocadherin 2 alpha b 3 |
| chr5_-_16983336 | 1.05 |
ENSDART00000038740
|
galnt9
|
polypeptide N-acetylgalactosaminyltransferase 9 |
| chr4_-_390431 | 1.04 |
ENSDART00000067482
ENSDART00000138500 |
dynlt1
|
dynein, light chain, Tctex-type 1 |
| chr1_+_53954230 | 1.04 |
ENSDART00000037729
ENSDART00000159900 |
ccsapa
|
centriole, cilia and spindle-associated protein a |
| chr3_-_41791178 | 1.04 |
ENSDART00000049687
|
grifin
|
galectin-related inter-fiber protein |
| chr10_-_14929392 | 1.03 |
ENSDART00000137430
|
smad2
|
SMAD family member 2 |
| chr1_-_30510839 | 1.03 |
ENSDART00000168189
ENSDART00000174868 |
igf2bp2b
|
insulin-like growth factor 2 mRNA binding protein 2b |
| chr24_-_39772045 | 1.01 |
ENSDART00000087441
|
GFOD1
|
si:ch211-276f18.2 |
| chr2_-_48171112 | 1.00 |
ENSDART00000156258
|
pfkpb
|
phosphofructokinase, platelet b |
| chr17_-_30975707 | 0.99 |
ENSDART00000138346
|
evla
|
Enah/Vasp-like a |
| chr6_+_36381709 | 0.99 |
ENSDART00000004727
|
rhcgl1
|
Rh family, C glycoprotein, like 1 |
| chr17_+_3993772 | 0.99 |
ENSDART00000170822
ENSDART00000168613 ENSDART00000186174 |
tmx4
|
thioredoxin-related transmembrane protein 4 |
| chr24_-_1985007 | 0.99 |
ENSDART00000189870
|
PARD3 (1 of many)
|
par-3 family cell polarity regulator |
| chr4_-_27398385 | 0.98 |
ENSDART00000142117
ENSDART00000150553 ENSDART00000182746 |
fam19a5a
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5a |
| chr19_+_935565 | 0.97 |
ENSDART00000113368
|
RNF5
|
ring finger protein 5 |
| chr8_+_52637507 | 0.96 |
ENSDART00000163830
|
si:dkey-90l8.3
|
si:dkey-90l8.3 |
| chr6_-_37469775 | 0.96 |
ENSDART00000156546
|
plcxd1
|
phosphatidylinositol-specific phospholipase C, X domain containing 1 |
| chr9_-_7212973 | 0.95 |
ENSDART00000133638
|
mgat4a
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr9_-_6380653 | 0.95 |
ENSDART00000078523
|
ecrg4a
|
esophageal cancer related gene 4a |
| chr21_-_37733287 | 0.94 |
ENSDART00000157826
|
mpp1
|
membrane protein, palmitoylated 1 |
| chr9_+_38481780 | 0.94 |
ENSDART00000087241
|
lss
|
lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) |
| chr7_-_58098814 | 0.93 |
ENSDART00000147287
ENSDART00000043984 |
ank2b
|
ankyrin 2b, neuronal |
| chr17_+_12075805 | 0.93 |
ENSDART00000155329
|
cnsta
|
consortin, connexin sorting protein a |
| chr25_-_4146947 | 0.93 |
ENSDART00000129268
|
fads2
|
fatty acid desaturase 2 |
| chr14_-_4121052 | 0.92 |
ENSDART00000167074
|
irf2
|
interferon regulatory factor 2 |
| chr6_-_6346557 | 0.92 |
ENSDART00000115018
|
ccdc88aa
|
coiled-coil domain containing 88Aa |
| chr17_+_3128273 | 0.92 |
ENSDART00000122453
|
zgc:136872
|
zgc:136872 |
| chr14_+_3495542 | 0.91 |
ENSDART00000168934
|
gstp2
|
glutathione S-transferase pi 2 |
| chr13_-_34683370 | 0.91 |
ENSDART00000113661
|
kif16bb
|
kinesin family member 16Bb |
| chr24_-_17892325 | 0.91 |
ENSDART00000154039
ENSDART00000185619 ENSDART00000178326 |
cntnap2a
|
contactin associated protein like 2a |
| chr17_+_42842632 | 0.90 |
ENSDART00000155547
ENSDART00000126087 |
qpct
|
glutaminyl-peptide cyclotransferase |
| chr14_-_33613794 | 0.89 |
ENSDART00000010022
|
xpnpep2
|
X-prolyl aminopeptidase (aminopeptidase P) 2, membrane-bound |
| chr10_-_42685512 | 0.88 |
ENSDART00000081347
|
stc1l
|
stanniocalcin 1, like |
| chr11_+_36409457 | 0.87 |
ENSDART00000077641
|
cyb561d1
|
cytochrome b561 family, member D1 |
| chr17_-_6076084 | 0.87 |
ENSDART00000058890
|
ephx2
|
epoxide hydrolase 2, cytoplasmic |
| chr8_+_28900689 | 0.87 |
ENSDART00000141634
|
grid2
|
glutamate receptor, ionotropic, delta 2 |
| chr24_+_5208171 | 0.87 |
ENSDART00000155926
ENSDART00000154464 |
si:ch73-206p6.1
|
si:ch73-206p6.1 |
| chr24_-_39771667 | 0.84 |
ENSDART00000181867
|
GFOD1
|
si:ch211-276f18.2 |
| chr1_-_44413629 | 0.84 |
ENSDART00000192747
|
CT583626.1
|
|
| chr5_-_18897482 | 0.83 |
ENSDART00000010101
ENSDART00000099434 |
aacs
|
acetoacetyl-CoA synthetase |
| chr13_+_19884631 | 0.83 |
ENSDART00000089533
|
atrnl1a
|
attractin-like 1a |
| chr18_+_16125852 | 0.83 |
ENSDART00000061106
|
bhlhe41
|
basic helix-loop-helix family, member e41 |
| chr6_-_24053404 | 0.82 |
ENSDART00000168511
|
si:dkey-44g17.6
|
si:dkey-44g17.6 |
| chr9_+_48943471 | 0.81 |
ENSDART00000125090
|
stk39
|
serine threonine kinase 39 |
| chr12_-_33706726 | 0.80 |
ENSDART00000153135
|
myo15b
|
myosin XVB |
| chr15_+_46356879 | 0.80 |
ENSDART00000154388
|
wu:fb18f06
|
wu:fb18f06 |
| chr22_+_38301365 | 0.79 |
ENSDART00000137339
|
traf5
|
Tnf receptor-associated factor 5 |
| chr8_-_49399470 | 0.79 |
ENSDART00000136395
|
si:ch211-194e1.7
|
si:ch211-194e1.7 |
| chr10_-_35103208 | 0.77 |
ENSDART00000192734
|
zgc:110006
|
zgc:110006 |
| chr3_-_23407720 | 0.77 |
ENSDART00000155658
|
rapgefl1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr11_+_30162407 | 0.77 |
ENSDART00000190333
ENSDART00000127502 |
cdkl5
|
cyclin-dependent kinase-like 5 |
| chr11_-_42918971 | 0.76 |
ENSDART00000052912
|
pcdh20
|
protocadherin 20 |
| chr5_-_16425781 | 0.76 |
ENSDART00000185624
ENSDART00000180617 |
slc39a14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr11_+_23760470 | 0.76 |
ENSDART00000175688
ENSDART00000121874 ENSDART00000086720 |
nfasca
|
neurofascin homolog (chicken) a |
| chr14_-_4044545 | 0.76 |
ENSDART00000169527
|
snx25
|
sorting nexin 25 |
| chr25_+_4787431 | 0.76 |
ENSDART00000170640
|
myo5c
|
myosin VC |
| chr19_-_5805923 | 0.75 |
ENSDART00000134340
|
si:ch211-264f5.8
|
si:ch211-264f5.8 |
| chr11_+_30161699 | 0.75 |
ENSDART00000190504
|
cdkl5
|
cyclin-dependent kinase-like 5 |
| chr12_+_17504559 | 0.74 |
ENSDART00000020628
|
cyth3a
|
cytohesin 3a |
| chr13_-_9895564 | 0.74 |
ENSDART00000169831
ENSDART00000142629 |
si:ch211-117n7.6
|
si:ch211-117n7.6 |
| chr20_+_22045089 | 0.74 |
ENSDART00000063564
ENSDART00000187013 ENSDART00000161552 ENSDART00000174478 ENSDART00000063568 ENSDART00000152247 |
nmu
|
neuromedin U |
| chr21_+_26071874 | 0.74 |
ENSDART00000003001
ENSDART00000146573 |
rpl23a
|
ribosomal protein L23a |
| chr10_+_2582254 | 0.74 |
ENSDART00000016103
|
nxnl2
|
nucleoredoxin like 2 |
| chr17_-_6076266 | 0.74 |
ENSDART00000171084
|
ephx2
|
epoxide hydrolase 2, cytoplasmic |
| chr20_-_3087162 | 0.73 |
ENSDART00000152495
|
map3k5
|
mitogen-activated protein kinase kinase kinase 5 |
| chr2_+_927204 | 0.73 |
ENSDART00000165477
|
AL935300.3
|
|
| chr5_+_45138934 | 0.73 |
ENSDART00000041412
ENSDART00000136002 |
smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr5_+_21931124 | 0.73 |
ENSDART00000137627
|
si:ch73-92i20.1
|
si:ch73-92i20.1 |
| chr20_-_1268863 | 0.72 |
ENSDART00000109321
ENSDART00000027119 |
lats1
|
large tumor suppressor kinase 1 |
| chr14_-_45967981 | 0.72 |
ENSDART00000188062
|
macrod1
|
MACRO domain containing 1 |
| chr6_+_27275716 | 0.71 |
ENSDART00000156434
ENSDART00000114347 |
scly
|
selenocysteine lyase |
| chr8_-_65189 | 0.71 |
ENSDART00000168412
|
hsd17b4
|
hydroxysteroid (17-beta) dehydrogenase 4 |
| chr13_+_47821524 | 0.71 |
ENSDART00000109978
|
zc3h6
|
zinc finger CCCH-type containing 6 |
| chr15_-_30815826 | 0.71 |
ENSDART00000156160
ENSDART00000145918 |
msi2b
|
musashi RNA-binding protein 2b |
| chr5_-_64355227 | 0.70 |
ENSDART00000170787
|
fam78aa
|
family with sequence similarity 78, member Aa |
| chr16_-_383664 | 0.69 |
ENSDART00000051693
|
irx4a
|
iroquois homeobox 4a |
| chr3_-_45848257 | 0.69 |
ENSDART00000147198
|
igfals
|
insulin-like growth factor binding protein, acid labile subunit |
| chr16_+_36650984 | 0.69 |
ENSDART00000156780
|
col6a4a
|
collagen, type VI, alpha 4a |
| chr18_-_35390735 | 0.68 |
ENSDART00000098297
|
CABZ01040999.1
|
|
| chr1_-_36152131 | 0.68 |
ENSDART00000182113
ENSDART00000182904 |
znf827
|
zinc finger protein 827 |
| chr9_+_747612 | 0.67 |
ENSDART00000181004
|
insig2
|
insulin induced gene 2 |
| chr11_+_25583950 | 0.67 |
ENSDART00000111961
|
ccdc120
|
coiled-coil domain containing 120 |
| chr21_-_3826706 | 0.67 |
ENSDART00000053607
|
st6galnac6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr1_-_56080112 | 0.66 |
ENSDART00000075469
ENSDART00000161473 |
c3a.6
|
complement component c3a, duplicate 6 |
| chr2_+_6926100 | 0.66 |
ENSDART00000153289
|
nos1apb
|
nitric oxide synthase 1 (neuronal) adaptor protein b |
| chr19_-_7450796 | 0.66 |
ENSDART00000104750
|
mllt11
|
MLLT11, transcription factor 7 cofactor |
| chr18_-_19405616 | 0.65 |
ENSDART00000191290
ENSDART00000090855 |
megf11
|
multiple EGF-like-domains 11 |
| chr3_+_49021079 | 0.64 |
ENSDART00000162012
|
zgc:163083
|
zgc:163083 |
| chr17_+_38476300 | 0.63 |
ENSDART00000123298
|
stard9
|
StAR-related lipid transfer (START) domain containing 9 |
| chr10_+_2669405 | 0.63 |
ENSDART00000110202
|
ccl27b
|
chemokine (C-C motif) ligand 27b |
| chr9_-_1702648 | 0.63 |
ENSDART00000102934
|
hnrnpa3
|
heterogeneous nuclear ribonucleoprotein A3 |
| chr11_-_37997419 | 0.63 |
ENSDART00000102870
|
slc41a1
|
solute carrier family 41 (magnesium transporter), member 1 |
| chr17_-_3190070 | 0.63 |
ENSDART00000115027
|
tmem151bb
|
transmembrane protein 151Bb |
| chr5_+_22177033 | 0.62 |
ENSDART00000131223
|
fdx1b
|
ferredoxin 1b |
| chr17_+_43623598 | 0.62 |
ENSDART00000154138
|
znf365
|
zinc finger protein 365 |
| chr5_+_64732036 | 0.62 |
ENSDART00000073950
|
olfm1a
|
olfactomedin 1a |
| chr6_-_30932078 | 0.62 |
ENSDART00000028612
|
pde4ba
|
phosphodiesterase 4B, cAMP-specific a |
| chr17_-_8570257 | 0.62 |
ENSDART00000154713
ENSDART00000121488 |
fzd3b
|
frizzled class receptor 3b |
| chr5_+_44228526 | 0.62 |
ENSDART00000143789
|
TMEM8B
|
si:dkey-84j12.1 |
| chr7_-_72427616 | 0.61 |
ENSDART00000181878
|
rph3ab
|
rabphilin 3A homolog (mouse), b |
| chr22_+_35275468 | 0.61 |
ENSDART00000189516
ENSDART00000181572 ENSDART00000165353 ENSDART00000185352 |
rubcn
|
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein |
| chr24_-_6898302 | 0.61 |
ENSDART00000158646
|
dpp6a
|
dipeptidyl-peptidase 6a |
| chr1_-_53756851 | 0.61 |
ENSDART00000122445
|
akt3b
|
v-akt murine thymoma viral oncogene homolog 3b |
| chr5_+_39504136 | 0.61 |
ENSDART00000121460
|
prdm8b
|
PR domain containing 8b |
| chr11_-_2478374 | 0.60 |
ENSDART00000173205
|
si:ch73-267c23.10
|
si:ch73-267c23.10 |
| chr2_-_23778180 | 0.60 |
ENSDART00000136782
|
si:dkey-24c2.7
|
si:dkey-24c2.7 |
| chr4_+_12292274 | 0.60 |
ENSDART00000061070
ENSDART00000150786 |
mkrn1
|
makorin, ring finger protein, 1 |
| chr14_+_15231097 | 0.60 |
ENSDART00000172430
|
si:dkey-203a12.3
|
si:dkey-203a12.3 |
| chr5_-_32274383 | 0.60 |
ENSDART00000122889
|
myhz1.3
|
myosin, heavy polypeptide 1.3, skeletal muscle |
| chr14_-_49133426 | 0.59 |
ENSDART00000042421
|
ppp2ca
|
protein phosphatase 2, catalytic subunit, alpha isozyme |
| chr7_-_69521481 | 0.59 |
ENSDART00000148465
|
slc1a1
|
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 |
| chr4_+_76426308 | 0.58 |
ENSDART00000180424
ENSDART00000159789 |
FP074874.1
zgc:172128
|
zgc:172128 |
| chr5_+_51848756 | 0.58 |
ENSDART00000087467
ENSDART00000184466 |
cmya5
|
cardiomyopathy associated 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of pou2f3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0097237 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.5 | 2.0 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.4 | 1.7 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.4 | 1.7 | GO:0099548 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.4 | 1.7 | GO:0009447 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.4 | 1.1 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.3 | 1.4 | GO:0016108 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 0.3 | 2.9 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.3 | 0.9 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.3 | 0.9 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.3 | 1.2 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.3 | 1.4 | GO:0050996 | positive regulation of lipid catabolic process(GO:0050996) |
| 0.3 | 1.1 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.3 | 0.8 | GO:0033212 | iron assimilation(GO:0033212) |
| 0.2 | 0.5 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.2 | 0.9 | GO:0018199 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) peptidyl-glutamine modification(GO:0018199) |
| 0.2 | 0.7 | GO:2000639 | response to sterol(GO:0036314) cellular response to sterol(GO:0036315) SREBP-SCAP complex retention in endoplasmic reticulum(GO:0036316) regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.2 | 2.5 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) |
| 0.2 | 1.1 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.2 | 0.6 | GO:0090219 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of lipid kinase activity(GO:0090219) |
| 0.2 | 2.0 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.2 | 1.4 | GO:0032475 | otolith formation(GO:0032475) |
| 0.2 | 0.9 | GO:0010447 | response to acidic pH(GO:0010447) negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.2 | 1.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.2 | 1.5 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.2 | 0.9 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.2 | 0.9 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.2 | 0.5 | GO:0051580 | regulation of neurotransmitter uptake(GO:0051580) negative regulation of anion transport(GO:1903792) |
| 0.2 | 1.9 | GO:0032196 | transposition(GO:0032196) |
| 0.2 | 0.5 | GO:0048677 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.2 | 0.5 | GO:0035046 | pronuclear migration(GO:0035046) |
| 0.1 | 0.7 | GO:0046462 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.1 | 1.0 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.1 | 1.6 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 0.7 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.1 | 1.0 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.1 | 0.7 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 7.4 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.1 | 0.6 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.1 | 1.4 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.1 | 0.9 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 0.1 | 0.8 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.1 | 0.5 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 | 1.0 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.1 | 1.0 | GO:0006735 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 0.5 | GO:1902047 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.1 | 0.3 | GO:0046125 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.1 | 0.2 | GO:0072526 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 0.1 | 0.3 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.7 | GO:0061075 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.1 | 1.2 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.1 | 0.4 | GO:0007508 | larval development(GO:0002164) larval heart development(GO:0007508) angiogenesis involved in wound healing(GO:0060055) |
| 0.1 | 0.8 | GO:0030431 | sleep(GO:0030431) |
| 0.1 | 0.2 | GO:0010142 | farnesyl diphosphate biosynthetic process, mevalonate pathway(GO:0010142) isoprenoid biosynthetic process via mevalonate(GO:1902767) |
| 0.1 | 1.2 | GO:0071545 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) inositol phosphate catabolic process(GO:0071545) |
| 0.1 | 1.6 | GO:0060841 | venous blood vessel development(GO:0060841) |
| 0.1 | 0.8 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 1.4 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.1 | 0.7 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.1 | 0.6 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.1 | 0.5 | GO:0016119 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.1 | 0.4 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 0.5 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.1 | 0.3 | GO:0016081 | synaptic vesicle docking(GO:0016081) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.1 | 2.3 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.2 | GO:0043618 | regulation of transcription from RNA polymerase II promoter in response to stress(GO:0043618) regulation of DNA-templated transcription in response to stress(GO:0043620) |
| 0.0 | 0.5 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.8 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.7 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.9 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.9 | GO:0016114 | terpenoid biosynthetic process(GO:0016114) |
| 0.0 | 0.7 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 1.2 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.6 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.3 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.4 | GO:0071450 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.7 | GO:0048923 | posterior lateral line neuromast hair cell differentiation(GO:0048923) |
| 0.0 | 5.2 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.8 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.3 | GO:0046294 | formaldehyde metabolic process(GO:0046292) formaldehyde catabolic process(GO:0046294) |
| 0.0 | 3.0 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.2 | GO:0070206 | protein trimerization(GO:0070206) protein homotrimerization(GO:0070207) |
| 0.0 | 1.4 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.3 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) synaptic transmission, GABAergic(GO:0051932) |
| 0.0 | 5.0 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 0.1 | GO:0060043 | regulation of cardiac muscle tissue growth(GO:0055021) regulation of cardiac muscle cell proliferation(GO:0060043) |
| 0.0 | 0.8 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.3 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.6 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.4 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.0 | 0.8 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.3 | GO:0006567 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.0 | 0.6 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.3 | GO:0019883 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.2 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.0 | 0.1 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.3 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.2 | GO:0046620 | regulation of organ growth(GO:0046620) |
| 0.0 | 0.2 | GO:0097241 | hematopoietic stem cell migration to bone marrow(GO:0097241) |
| 0.0 | 0.6 | GO:0042181 | ketone biosynthetic process(GO:0042181) |
| 0.0 | 0.4 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.4 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.5 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 1.5 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.7 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.6 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.9 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.2 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.6 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.0 | 1.3 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 4.1 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 0.2 | GO:0034434 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.0 | 1.2 | GO:0071482 | cellular response to light stimulus(GO:0071482) |
| 0.0 | 0.8 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.3 | GO:0002027 | regulation of heart rate(GO:0002027) |
| 0.0 | 0.7 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.8 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
| 0.0 | 0.6 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 0.9 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.9 | GO:0051592 | response to calcium ion(GO:0051592) |
| 0.0 | 1.2 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 0.4 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.4 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.5 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 1.3 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.5 | GO:0035567 | non-canonical Wnt signaling pathway(GO:0035567) |
| 0.0 | 0.6 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.2 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.0 | 0.7 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.2 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 1.4 | GO:0031098 | stress-activated protein kinase signaling cascade(GO:0031098) |
| 0.0 | 0.7 | GO:0043280 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043280) |
| 0.0 | 0.1 | GO:0046473 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.2 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.3 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.6 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.7 | GO:0006956 | complement activation(GO:0006956) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.2 | 0.7 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.2 | 1.5 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.2 | 7.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.7 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.6 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 0.9 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.1 | 1.8 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 0.4 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.1 | 7.4 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 0.4 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 1.0 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 1.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 3.0 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 0.3 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 0.9 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 1.1 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 6.0 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 1.7 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 1.1 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.7 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.5 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.6 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 2.5 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 1.2 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 1.0 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 1.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 1.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 5.0 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 2.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.6 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 3.2 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 2.1 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 1.6 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.2 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.4 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.2 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.0 | GO:0005521 | lamin binding(GO:0005521) |
| 0.5 | 1.6 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.5 | 2.0 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.5 | 1.4 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.5 | 1.4 | GO:0005131 | growth hormone receptor binding(GO:0005131) growth hormone activity(GO:0070186) |
| 0.4 | 1.1 | GO:0052833 | inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.3 | 1.0 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.3 | 1.4 | GO:0052885 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.3 | 1.7 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.3 | 0.8 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.2 | 1.7 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.2 | 0.9 | GO:0071253 | connexin binding(GO:0071253) |
| 0.2 | 0.9 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.2 | 0.6 | GO:0030882 | lipid antigen binding(GO:0030882) |
| 0.2 | 1.5 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 0.5 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.2 | 0.5 | GO:0005252 | open rectifier potassium channel activity(GO:0005252) |
| 0.2 | 1.7 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.2 | 4.5 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 0.5 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 1.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.7 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.1 | 0.7 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 0.8 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 1.7 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 1.0 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 0.5 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.1 | 3.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 2.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.4 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.1 | 1.2 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.1 | 0.9 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.3 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 1.1 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.1 | 0.3 | GO:0047611 | tubulin deacetylase activity(GO:0042903) acetylspermidine deacetylase activity(GO:0047611) |
| 0.1 | 0.2 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.1 | 0.4 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 4.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.7 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.7 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 0.4 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 1.4 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.1 | 0.5 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.1 | 1.1 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 1.6 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 1.2 | GO:0008252 | nucleotidase activity(GO:0008252) |
| 0.1 | 0.5 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.1 | 0.3 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) S-(hydroxymethyl)glutathione dehydrogenase activity(GO:0051903) |
| 0.1 | 0.3 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.1 | 0.3 | GO:0002058 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.0 | 1.1 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.9 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.2 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.2 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 1.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.3 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 1.4 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 1.8 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.5 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 1.1 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.6 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.4 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.1 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.0 | 0.3 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 0.2 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.0 | 0.2 | GO:0034736 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.0 | 2.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 4.8 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 1.0 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 3.2 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.6 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.7 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 1.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.8 | GO:0016878 | acid-thiol ligase activity(GO:0016878) |
| 0.0 | 0.5 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.5 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.4 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.9 | GO:0016866 | intramolecular transferase activity(GO:0016866) |
| 0.0 | 0.4 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.9 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.0 | 0.9 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.3 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.3 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.4 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 0.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.6 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 3.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.8 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.7 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.3 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.0 | 0.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.9 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.5 | GO:0016849 | phosphorus-oxygen lyase activity(GO:0016849) |
| 0.0 | 0.4 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 2.6 | GO:0042277 | peptide binding(GO:0042277) |
| 0.0 | 0.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 1.4 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 1.7 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 0.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.6 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 2.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 1.1 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 1.6 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 1.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 1.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.7 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.1 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.7 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.4 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.5 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.2 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.5 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.2 | 3.6 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.2 | 2.8 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.2 | 1.6 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 1.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 0.6 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 1.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.6 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 1.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 0.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 1.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.7 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 1.0 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.1 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 1.1 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.6 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.9 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |
| 0.0 | 0.9 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.4 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.4 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.1 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.2 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.7 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |