Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
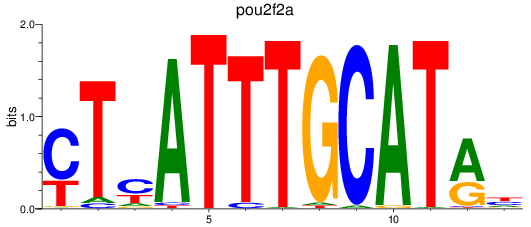
Results for pou2f2a
Z-value: 1.07
Transcription factors associated with pou2f2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pou2f2a
|
ENSDARG00000019658 | POU class 2 homeobox 2a |
|
pou2f2a
|
ENSDARG00000036816 | POU class 2 homeobox 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pou2f2a | dr11_v1_chr16_+_10918252_10918252 | -0.66 | 2.1e-03 | Click! |
Activity profile of pou2f2a motif
Sorted Z-values of pou2f2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_6081803 | 4.78 |
ENSDART00000099224
|
dld
|
deltaD |
| chr23_+_21473103 | 4.67 |
ENSDART00000142921
|
si:ch73-21g5.7
|
si:ch73-21g5.7 |
| chr25_+_22320738 | 3.56 |
ENSDART00000073566
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr4_+_15968483 | 3.33 |
ENSDART00000101575
|
si:dkey-117n7.5
|
si:dkey-117n7.5 |
| chr14_-_32965169 | 3.07 |
ENSDART00000114973
|
cdx4
|
caudal type homeobox 4 |
| chr11_-_11575070 | 2.62 |
ENSDART00000142208
|
zgc:110712
|
zgc:110712 |
| chr14_-_26704829 | 2.38 |
ENSDART00000078563
|
neurog1
|
neurogenin 1 |
| chr5_-_66028371 | 2.28 |
ENSDART00000183012
|
nrarpb
|
NOTCH regulated ankyrin repeat protein b |
| chr7_+_15736230 | 2.11 |
ENSDART00000109942
|
mctp2b
|
multiple C2 domains, transmembrane 2b |
| chr23_+_7692042 | 2.05 |
ENSDART00000018512
|
pofut1
|
protein O-fucosyltransferase 1 |
| chr23_+_12454542 | 2.04 |
ENSDART00000182259
ENSDART00000193043 |
si:ch211-153a8.4
|
si:ch211-153a8.4 |
| chr5_-_66028714 | 1.87 |
ENSDART00000022625
ENSDART00000164228 |
nrarpb
|
NOTCH regulated ankyrin repeat protein b |
| chr17_-_4245311 | 1.84 |
ENSDART00000055379
|
gdf3
|
growth differentiation factor 3 |
| chr15_+_7187228 | 1.75 |
ENSDART00000109394
|
her13
|
hairy-related 13 |
| chr20_+_48782068 | 1.71 |
ENSDART00000159275
|
nkx2.2b
|
NK2 homeobox 2b |
| chr1_+_54013457 | 1.67 |
ENSDART00000012104
ENSDART00000126339 |
dla
|
deltaA |
| chr1_-_51720633 | 1.59 |
ENSDART00000045894
|
rnaseh2a
|
ribonuclease H2, subunit A |
| chr21_-_34926619 | 1.56 |
ENSDART00000065337
ENSDART00000192740 |
kif20a
|
kinesin family member 20A |
| chr21_-_44512893 | 1.54 |
ENSDART00000166853
|
zgc:136410
|
zgc:136410 |
| chr22_+_35930526 | 1.54 |
ENSDART00000169242
|
LO016987.1
|
|
| chr19_+_19775757 | 1.51 |
ENSDART00000164677
|
hoxa3a
|
homeobox A3a |
| chr23_+_23232136 | 1.47 |
ENSDART00000126479
ENSDART00000187764 |
plekhn1
|
pleckstrin homology domain containing, family N member 1 |
| chr14_+_35383267 | 1.46 |
ENSDART00000143818
|
clint1a
|
clathrin interactor 1a |
| chr4_-_9780931 | 1.44 |
ENSDART00000134280
ENSDART00000150664 ENSDART00000150304 ENSDART00000080744 |
svopl
|
SVOP-like |
| chr19_-_41518922 | 1.41 |
ENSDART00000164483
ENSDART00000062080 |
chrac1
|
chromatin accessibility complex 1 |
| chr5_-_65662996 | 1.40 |
ENSDART00000147707
|
camsap1b
|
calmodulin regulated spectrin-associated protein 1b |
| chr11_-_30158609 | 1.36 |
ENSDART00000006669
|
scml2
|
Scm polycomb group protein like 2 |
| chr17_+_23311377 | 1.35 |
ENSDART00000128073
|
ppp1r3ca
|
protein phosphatase 1, regulatory subunit 3Ca |
| chr10_+_10351685 | 1.33 |
ENSDART00000109432
|
cercam
|
cerebral endothelial cell adhesion molecule |
| chr8_+_28593707 | 1.33 |
ENSDART00000097213
|
tcf15
|
transcription factor 15 |
| chr10_-_7555326 | 1.32 |
ENSDART00000162191
ENSDART00000186945 |
wrn
|
Werner syndrome |
| chr13_-_30996072 | 1.32 |
ENSDART00000181661
|
wdfy4
|
WDFY family member 4 |
| chr3_-_5964557 | 1.31 |
ENSDART00000184738
|
BX284638.1
|
|
| chr2_+_4146606 | 1.31 |
ENSDART00000171170
|
mib1
|
mindbomb E3 ubiquitin protein ligase 1 |
| chr19_+_9459050 | 1.29 |
ENSDART00000186419
|
si:ch211-288g17.3
|
si:ch211-288g17.3 |
| chr12_+_16087077 | 1.28 |
ENSDART00000141898
|
znf281b
|
zinc finger protein 281b |
| chr4_-_64141714 | 1.28 |
ENSDART00000128628
|
BX914205.3
|
|
| chr2_+_4146299 | 1.27 |
ENSDART00000173418
|
mib1
|
mindbomb E3 ubiquitin protein ligase 1 |
| chr19_-_18152942 | 1.23 |
ENSDART00000190182
|
nfe2l3
|
nuclear factor, erythroid 2-like 3 |
| chr14_-_38929885 | 1.20 |
ENSDART00000148737
|
btk
|
Bruton agammaglobulinemia tyrosine kinase |
| chr7_-_32020858 | 1.20 |
ENSDART00000023568
|
kif18a
|
kinesin family member 18A |
| chr1_+_24557414 | 1.19 |
ENSDART00000076519
|
dctpp1
|
dCTP pyrophosphatase 1 |
| chr7_+_20917966 | 1.18 |
ENSDART00000129161
|
wrap53
|
WD repeat containing, antisense to TP53 |
| chr10_-_1961930 | 1.17 |
ENSDART00000122446
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr23_-_1017428 | 1.17 |
ENSDART00000110588
ENSDART00000183158 |
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr9_+_29431763 | 1.17 |
ENSDART00000186095
ENSDART00000182640 |
uggt2
|
UDP-glucose glycoprotein glucosyltransferase 2 |
| chr20_+_53441935 | 1.17 |
ENSDART00000175214
|
apobb.2
|
apolipoprotein Bb, tandem duplicate 2 |
| chr11_+_25010491 | 1.16 |
ENSDART00000167285
|
zgc:92107
|
zgc:92107 |
| chr4_+_58695514 | 1.14 |
ENSDART00000169840
|
si:dkey-25i10.1
|
si:dkey-25i10.1 |
| chr1_-_46832880 | 1.13 |
ENSDART00000142406
|
si:ch73-160h15.3
|
si:ch73-160h15.3 |
| chr7_-_41512999 | 1.13 |
ENSDART00000173577
|
si:dkey-10f23.2
|
si:dkey-10f23.2 |
| chr4_+_29206813 | 1.13 |
ENSDART00000131893
|
si:dkey-23a23.1
|
si:dkey-23a23.1 |
| chr23_-_1017605 | 1.12 |
ENSDART00000138290
|
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr17_-_4245902 | 1.12 |
ENSDART00000151851
|
gdf3
|
growth differentiation factor 3 |
| chr6_-_9581949 | 1.12 |
ENSDART00000144335
|
cyp27c1
|
cytochrome P450, family 27, subfamily C, polypeptide 1 |
| chr16_+_25074029 | 1.11 |
ENSDART00000155465
|
si:dkeyp-84f3.9
|
si:dkeyp-84f3.9 |
| chr4_+_9592486 | 1.11 |
ENSDART00000080829
|
hspa14
|
heat shock protein 14 |
| chr16_+_38167883 | 1.10 |
ENSDART00000111796
|
pi4kb
|
phosphatidylinositol 4-kinase, catalytic, beta |
| chr12_+_23424108 | 1.10 |
ENSDART00000077732
|
bambia
|
BMP and activin membrane-bound inhibitor (Xenopus laevis) homolog a |
| chr5_-_63302944 | 1.10 |
ENSDART00000047110
|
gsnb
|
gelsolin b |
| chr7_-_57637779 | 1.10 |
ENSDART00000028017
|
mad2l1
|
MAD2 mitotic arrest deficient-like 1 (yeast) |
| chr24_-_23758003 | 1.10 |
ENSDART00000178085
|
BX640462.2
|
Danio rerio minichromosome maintenance domain containing 2 (mcmdc2), mRNA. |
| chr9_-_27398369 | 1.09 |
ENSDART00000186499
|
tex30
|
testis expressed 30 |
| chr4_+_64549970 | 1.07 |
ENSDART00000167846
|
si:ch211-223a21.3
|
si:ch211-223a21.3 |
| chr3_-_27646070 | 1.06 |
ENSDART00000122031
ENSDART00000151027 |
si:ch211-157c3.4
|
si:ch211-157c3.4 |
| chr18_+_22174630 | 1.05 |
ENSDART00000089549
|
ripor1
|
RHO family interacting cell polarization regulator 1 |
| chr12_+_27096835 | 1.05 |
ENSDART00000149475
|
ttll6
|
tubulin tyrosine ligase-like family, member 6 |
| chr16_-_21912210 | 1.04 |
ENSDART00000165824
|
setdb1b
|
SET domain, bifurcated 1b |
| chr9_-_53666031 | 1.04 |
ENSDART00000126314
|
pcdh8
|
protocadherin 8 |
| chr19_-_18152407 | 1.04 |
ENSDART00000193264
ENSDART00000016135 |
nfe2l3
|
nuclear factor, erythroid 2-like 3 |
| chr24_-_31904924 | 1.02 |
ENSDART00000156060
ENSDART00000129741 ENSDART00000154276 |
si:ch73-78o10.1
|
si:ch73-78o10.1 |
| chr16_-_21620947 | 1.02 |
ENSDART00000115011
ENSDART00000183125 ENSDART00000188856 ENSDART00000189460 |
ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr16_-_31790285 | 1.01 |
ENSDART00000184655
|
chd4b
|
chromodomain helicase DNA binding protein 4b |
| chr1_-_25438737 | 1.01 |
ENSDART00000134470
|
fhdc1
|
FH2 domain containing 1 |
| chr5_+_63302660 | 0.99 |
ENSDART00000142131
|
si:ch73-376l24.2
|
si:ch73-376l24.2 |
| chr21_+_20903244 | 0.98 |
ENSDART00000186193
|
c7b
|
complement component 7b |
| chr7_-_8315179 | 0.95 |
ENSDART00000184049
|
f13a1b
|
coagulation factor XIII, A1 polypeptide b |
| chr5_+_45322734 | 0.94 |
ENSDART00000084411
|
adamts3
|
ADAM metallopeptidase with thrombospondin type 1 motif, 3 |
| chr3_-_26806032 | 0.94 |
ENSDART00000143710
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr12_-_35582683 | 0.93 |
ENSDART00000167933
|
sec24c
|
SEC24 homolog C, COPII coat complex component |
| chr13_+_48617974 | 0.92 |
ENSDART00000168596
|
si:ch1073-268j14.1
|
si:ch1073-268j14.1 |
| chr15_-_29388012 | 0.92 |
ENSDART00000115032
|
serpinh1b
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1b |
| chr2_-_22993601 | 0.91 |
ENSDART00000125049
ENSDART00000099735 |
mllt1b
|
MLLT1, super elongation complex subunit b |
| chr7_-_12065668 | 0.90 |
ENSDART00000101537
|
mex3b
|
mex-3 RNA binding family member B |
| chr5_+_9259971 | 0.89 |
ENSDART00000163060
|
susd1
|
sushi domain containing 1 |
| chr17_+_26965351 | 0.89 |
ENSDART00000114215
ENSDART00000147192 |
grhl3
|
grainyhead-like transcription factor 3 |
| chr23_+_16864130 | 0.89 |
ENSDART00000060181
|
zgc:114174
|
zgc:114174 |
| chr24_+_37825634 | 0.88 |
ENSDART00000129889
|
ift140
|
intraflagellar transport 140 homolog (Chlamydomonas) |
| chr18_+_30998472 | 0.87 |
ENSDART00000154993
ENSDART00000099333 |
cd151l
|
CD151 antigen, like |
| chr8_+_36570508 | 0.87 |
ENSDART00000145868
|
pold2
|
polymerase (DNA directed), delta 2, regulatory subunit |
| chr20_+_25625872 | 0.87 |
ENSDART00000078385
|
ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr23_-_20126257 | 0.87 |
ENSDART00000005021
|
tktb
|
transketolase b |
| chr22_+_5135884 | 0.86 |
ENSDART00000141276
|
mydgf
|
myeloid-derived growth factor |
| chr21_+_19070921 | 0.84 |
ENSDART00000029874
|
nkx6.1
|
NK6 homeobox 1 |
| chr18_+_6866276 | 0.84 |
ENSDART00000187516
|
dnaja2l
|
DnaJ (Hsp40) homolog, subfamily A, member 2, like |
| chr18_+_34667486 | 0.84 |
ENSDART00000163583
|
CR626874.1
|
|
| chr4_-_73351733 | 0.84 |
ENSDART00000169597
|
si:cabz01021426.2
|
si:cabz01021426.2 |
| chr23_+_12455295 | 0.83 |
ENSDART00000143752
ENSDART00000135785 |
si:ch211-153a8.4
|
si:ch211-153a8.4 |
| chr17_-_31579715 | 0.83 |
ENSDART00000110167
ENSDART00000191092 |
rpap1
|
RNA polymerase II associated protein 1 |
| chr10_-_1961576 | 0.82 |
ENSDART00000042441
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr2_+_56213694 | 0.82 |
ENSDART00000162582
|
upf1
|
upf1 regulator of nonsense transcripts homolog (yeast) |
| chr3_-_33422738 | 0.82 |
ENSDART00000075493
|
ccdc103
|
coiled-coil domain containing 103 |
| chr22_-_17611742 | 0.81 |
ENSDART00000144031
|
gpx4a
|
glutathione peroxidase 4a |
| chr22_+_2111820 | 0.81 |
ENSDART00000135500
|
znf1144
|
zinc finger protein 1144 |
| chr13_-_10431476 | 0.81 |
ENSDART00000133968
|
camkmt
|
calmodulin-lysine N-methyltransferase |
| chr15_-_16012963 | 0.81 |
ENSDART00000144138
|
hnf1ba
|
HNF1 homeobox Ba |
| chr25_+_5288665 | 0.80 |
ENSDART00000169540
|
CABZ01039861.1
|
|
| chr1_-_25438934 | 0.79 |
ENSDART00000111686
|
fhdc1
|
FH2 domain containing 1 |
| chr10_-_74408 | 0.79 |
ENSDART00000100073
ENSDART00000141723 |
dyrk1aa
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A, a |
| chr4_+_23126558 | 0.78 |
ENSDART00000162859
|
mdm2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr19_+_20178978 | 0.78 |
ENSDART00000145115
ENSDART00000151175 |
tra2a
|
transformer 2 alpha homolog |
| chr6_-_40286633 | 0.77 |
ENSDART00000033844
|
col7a1
|
collagen, type VII, alpha 1 |
| chr5_-_25072607 | 0.77 |
ENSDART00000145061
|
pnpla7b
|
patatin-like phospholipase domain containing 7b |
| chr9_+_22634073 | 0.76 |
ENSDART00000181822
|
etv5a
|
ets variant 5a |
| chr12_-_34887943 | 0.76 |
ENSDART00000027379
|
bicral
|
BRD4 interacting chromatin remodeling complex associated protein like |
| chr4_+_47656992 | 0.75 |
ENSDART00000161148
|
znf1040
|
zinc finger protein 1040 |
| chr4_+_59362813 | 0.75 |
ENSDART00000150655
|
znf1050
|
zinc finger protein 1050 |
| chr2_-_38206331 | 0.74 |
ENSDART00000136082
|
acin1a
|
apoptotic chromatin condensation inducer 1a |
| chr21_-_2185600 | 0.74 |
ENSDART00000169897
|
zgc:171220
|
zgc:171220 |
| chr21_+_26522571 | 0.71 |
ENSDART00000134617
|
adssl
|
adenylosuccinate synthase, like |
| chr8_-_39654669 | 0.71 |
ENSDART00000145677
|
si:dkey-63d15.12
|
si:dkey-63d15.12 |
| chr22_+_2364835 | 0.70 |
ENSDART00000174974
|
si:dkey-4c15.13
|
si:dkey-4c15.13 |
| chr20_-_22476255 | 0.69 |
ENSDART00000103510
|
pdgfra
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr15_-_29387446 | 0.69 |
ENSDART00000145976
ENSDART00000035096 |
serpinh1b
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1b |
| chr10_+_3145707 | 0.68 |
ENSDART00000160046
|
hic2
|
hypermethylated in cancer 2 |
| chr9_-_30502010 | 0.68 |
ENSDART00000149483
|
si:dkey-229b18.3
|
si:dkey-229b18.3 |
| chr11_+_19433936 | 0.68 |
ENSDART00000162081
|
prickle2b
|
prickle homolog 2b |
| chr17_+_24722646 | 0.68 |
ENSDART00000138356
|
mtfr1l
|
mitochondrial fission regulator 1-like |
| chr16_-_25380903 | 0.67 |
ENSDART00000086375
ENSDART00000188587 |
adnp2a
|
ADNP homeobox 2a |
| chr11_-_35575791 | 0.67 |
ENSDART00000031441
ENSDART00000188513 ENSDART00000183609 |
sema3fb
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Fb |
| chr1_-_51038885 | 0.67 |
ENSDART00000035150
|
spast
|
spastin |
| chr6_-_54111928 | 0.67 |
ENSDART00000083880
|
hyal2a
|
hyaluronoglucosaminidase 2a |
| chr6_+_41446541 | 0.66 |
ENSDART00000029553
ENSDART00000128756 ENSDART00000144864 |
rsg1
|
REM2 and RAB-like small GTPase 1 |
| chr15_-_34658057 | 0.66 |
ENSDART00000110964
|
bag6
|
BCL2 associated athanogene 6 |
| chr8_+_7315625 | 0.66 |
ENSDART00000135655
ENSDART00000181048 |
selenoh
|
selenoprotein H |
| chr1_-_55166511 | 0.65 |
ENSDART00000150430
ENSDART00000035725 |
pane1
|
proliferation associated nuclear element |
| chr5_+_33287611 | 0.65 |
ENSDART00000125093
ENSDART00000146759 |
med22
|
mediator complex subunit 22 |
| chr5_+_29726428 | 0.65 |
ENSDART00000143183
|
ddx31
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 31 |
| chr7_-_51461649 | 0.65 |
ENSDART00000193947
ENSDART00000174328 |
arhgap36
|
Rho GTPase activating protein 36 |
| chr2_-_7185460 | 0.65 |
ENSDART00000092078
|
rc3h1b
|
ring finger and CCCH-type domains 1b |
| chr5_+_38837429 | 0.65 |
ENSDART00000160236
|
fras1
|
Fraser extracellular matrix complex subunit 1 |
| chr4_-_48636872 | 0.64 |
ENSDART00000168605
|
znf1063
|
zinc finger protein 1063 |
| chr5_+_59494079 | 0.64 |
ENSDART00000148727
|
gtf2ird1
|
GTF2I repeat domain containing 1 |
| chr4_+_32649955 | 0.64 |
ENSDART00000135750
|
znf1042
|
zinc finger protein 1042 |
| chr8_+_36570791 | 0.64 |
ENSDART00000145566
ENSDART00000180527 |
pold2
|
polymerase (DNA directed), delta 2, regulatory subunit |
| chr12_+_4160804 | 0.63 |
ENSDART00000152515
|
itgam
|
integrin, alpha M (complement component 3 receptor 3 subunit) |
| chr24_+_35827766 | 0.63 |
ENSDART00000144700
|
si:dkeyp-7a3.1
|
si:dkeyp-7a3.1 |
| chr15_-_18574716 | 0.63 |
ENSDART00000142010
ENSDART00000019006 |
ncam1b
|
neural cell adhesion molecule 1b |
| chr21_-_2185004 | 0.63 |
ENSDART00000163405
|
zgc:171220
|
zgc:171220 |
| chr15_-_25083200 | 0.63 |
ENSDART00000156053
|
pimr191
|
Pim proto-oncogene, serine/threonine kinase, related 191 |
| chr19_-_21716593 | 0.62 |
ENSDART00000155126
|
znf516
|
zinc finger protein 516 |
| chr14_+_35428152 | 0.61 |
ENSDART00000172597
|
sytl4
|
synaptotagmin-like 4 |
| chr19_+_3140313 | 0.61 |
ENSDART00000125504
|
zgc:86598
|
zgc:86598 |
| chr4_-_56157199 | 0.60 |
ENSDART00000169806
|
si:ch211-207e19.15
|
si:ch211-207e19.15 |
| chr4_-_45337903 | 0.60 |
ENSDART00000159767
|
si:dkey-247i3.6
|
si:dkey-247i3.6 |
| chr6_+_56147812 | 0.60 |
ENSDART00000150219
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr3_-_15119856 | 0.60 |
ENSDART00000138328
|
xpo6
|
exportin 6 |
| chr20_+_25626479 | 0.60 |
ENSDART00000143883
|
ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr3_+_24275766 | 0.60 |
ENSDART00000055607
|
pdgfbb
|
platelet-derived growth factor beta polypeptide b |
| chr22_+_13917311 | 0.60 |
ENSDART00000022654
|
sh3bp4a
|
SH3-domain binding protein 4a |
| chr5_-_68782641 | 0.59 |
ENSDART00000141699
|
mepce
|
methylphosphate capping enzyme |
| chr4_-_35989745 | 0.59 |
ENSDART00000162568
|
znf1125
|
zinc finger protein 1125 |
| chr4_-_5831522 | 0.59 |
ENSDART00000008898
|
foxm1
|
forkhead box M1 |
| chr21_-_32467099 | 0.59 |
ENSDART00000186354
|
zgc:123105
|
zgc:123105 |
| chr6_-_22146258 | 0.58 |
ENSDART00000181700
|
gga1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1 |
| chr11_+_18612421 | 0.58 |
ENSDART00000110621
|
ncoa3
|
nuclear receptor coactivator 3 |
| chr3_-_58165254 | 0.57 |
ENSDART00000093031
|
snu13a
|
SNU13 homolog, small nuclear ribonucleoprotein a (U4/U6.U5) |
| chr24_+_29912509 | 0.57 |
ENSDART00000168422
|
frrs1b
|
ferric-chelate reductase 1b |
| chr1_+_31725154 | 0.57 |
ENSDART00000112333
ENSDART00000189801 |
cnnm2b
|
cyclin and CBS domain divalent metal cation transport mediator 2b |
| chr3_+_28581397 | 0.57 |
ENSDART00000150893
|
sept12
|
septin 12 |
| chr19_+_3653976 | 0.56 |
ENSDART00000125673
|
nedd9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr4_-_50434519 | 0.56 |
ENSDART00000150372
|
znf1061
|
zinc finger protein 1061 |
| chr23_+_2825940 | 0.56 |
ENSDART00000135781
|
plcg1
|
phospholipase C, gamma 1 |
| chr22_+_5135193 | 0.56 |
ENSDART00000106156
|
mydgf
|
myeloid-derived growth factor |
| chr1_-_54100988 | 0.55 |
ENSDART00000192662
|
rfx1b
|
regulatory factor X, 1b (influences HLA class II expression) |
| chr13_-_40411908 | 0.55 |
ENSDART00000057094
ENSDART00000150091 |
nkx2.3
|
NK2 homeobox 3 |
| chr19_+_19759577 | 0.55 |
ENSDART00000169480
|
hoxa5a
|
homeobox A5a |
| chr11_+_18612166 | 0.55 |
ENSDART00000162694
|
ncoa3
|
nuclear receptor coactivator 3 |
| chr11_-_30158191 | 0.54 |
ENSDART00000155278
ENSDART00000156121 |
scml2
|
Scm polycomb group protein like 2 |
| chr4_-_52621232 | 0.54 |
ENSDART00000124451
|
si:dkeyp-104f11.6
|
si:dkeyp-104f11.6 |
| chr18_-_14734678 | 0.54 |
ENSDART00000142462
|
tshz3a
|
teashirt zinc finger homeobox 3a |
| chr4_+_18489207 | 0.53 |
ENSDART00000135276
|
si:dkey-202b22.5
|
si:dkey-202b22.5 |
| chr5_+_26212621 | 0.53 |
ENSDART00000134432
|
oclnb
|
occludin b |
| chr22_-_10397600 | 0.53 |
ENSDART00000181964
ENSDART00000142886 |
nisch
|
nischarin |
| chr14_-_40389699 | 0.52 |
ENSDART00000181581
ENSDART00000173398 |
pcdh19
|
protocadherin 19 |
| chr1_+_7679328 | 0.52 |
ENSDART00000163488
ENSDART00000190070 |
en1b
|
engrailed homeobox 1b |
| chr8_-_24803111 | 0.52 |
ENSDART00000186281
|
BX324132.3
|
|
| chr4_+_31259 | 0.52 |
ENSDART00000166826
|
phtf2
|
putative homeodomain transcription factor 2 |
| chr12_-_33579873 | 0.52 |
ENSDART00000184661
|
tdrkh
|
tudor and KH domain containing |
| chr7_+_36041509 | 0.50 |
ENSDART00000162850
|
irx3a
|
iroquois homeobox 3a |
| chr4_+_33420247 | 0.50 |
ENSDART00000150868
|
znf1065
|
zinc finger protein 1065 |
| chr20_+_27464721 | 0.50 |
ENSDART00000189552
|
kif26aa
|
kinesin family member 26Aa |
| chr4_+_50726871 | 0.49 |
ENSDART00000150469
|
znf1058
|
zinc finger protein 1058 |
| chr2_-_17393971 | 0.49 |
ENSDART00000100201
|
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr19_+_9232676 | 0.49 |
ENSDART00000136957
|
kmt2ba
|
lysine (K)-specific methyltransferase 2Ba |
| chr20_-_25626693 | 0.48 |
ENSDART00000132247
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr4_-_67712607 | 0.48 |
ENSDART00000162955
|
znf1091
|
zinc finger protein 1091 |
| chr21_-_41070182 | 0.48 |
ENSDART00000026064
|
larsb
|
leucyl-tRNA synthetase b |
| chr6_+_33885828 | 0.48 |
ENSDART00000179994
|
gpbp1l1
|
GC-rich promoter binding protein 1-like 1 |
| chr19_+_43256986 | 0.47 |
ENSDART00000182336
|
DGKD
|
diacylglycerol kinase delta |
Network of associatons between targets according to the STRING database.
First level regulatory network of pou2f2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0007440 | foregut morphogenesis(GO:0007440) |
| 1.0 | 5.0 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.7 | 3.6 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.7 | 4.8 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.7 | 0.7 | GO:0031112 | positive regulation of microtubule polymerization or depolymerization(GO:0031112) |
| 0.7 | 2.0 | GO:0090008 | hypoblast development(GO:0090008) |
| 0.6 | 3.0 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.4 | 1.7 | GO:0055016 | hypochord development(GO:0055016) |
| 0.4 | 1.2 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.4 | 1.5 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.3 | 2.0 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.3 | 1.3 | GO:0044806 | multicellular organism aging(GO:0010259) G-quadruplex DNA unwinding(GO:0044806) |
| 0.3 | 0.6 | GO:0035477 | regulation of angioblast cell migration involved in selective angioblast sprouting(GO:0035477) |
| 0.3 | 0.8 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.3 | 1.6 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.3 | 1.8 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.2 | 1.2 | GO:0030576 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.2 | 1.4 | GO:0031113 | regulation of microtubule polymerization(GO:0031113) |
| 0.2 | 1.2 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.2 | 2.7 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.2 | 0.6 | GO:1903392 | epicardial cell to mesenchymal cell transition(GO:0003347) negative regulation of adherens junction organization(GO:1903392) |
| 0.2 | 0.6 | GO:1904871 | positive regulation of protein localization to nucleus(GO:1900182) protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.2 | 2.0 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.2 | 0.8 | GO:0048566 | embryonic digestive tract development(GO:0048566) |
| 0.2 | 0.5 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) protein localization to nuclear envelope(GO:0090435) |
| 0.2 | 0.5 | GO:0045649 | regulation of macrophage differentiation(GO:0045649) |
| 0.2 | 0.8 | GO:1904036 | negative regulation of epithelial cell apoptotic process(GO:1904036) |
| 0.1 | 0.6 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.1 | 1.7 | GO:0033505 | floor plate formation(GO:0021508) floor plate morphogenesis(GO:0033505) |
| 0.1 | 0.4 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.1 | 0.7 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.1 | 0.4 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.1 | 0.4 | GO:0061178 | insulin secretion involved in cellular response to glucose stimulus(GO:0035773) regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.1 | 0.4 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.1 | 0.6 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 1.4 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 0.4 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 0.4 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 1.4 | GO:0031937 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 1.5 | GO:0045682 | regulation of epidermis development(GO:0045682) |
| 0.1 | 1.5 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.1 | 0.2 | GO:0061323 | cell proliferation involved in heart morphogenesis(GO:0061323) regulation of cell proliferation involved in heart morphogenesis(GO:2000136) |
| 0.1 | 0.5 | GO:1902019 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.1 | 0.7 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.1 | 1.4 | GO:0001938 | positive regulation of endothelial cell proliferation(GO:0001938) |
| 0.1 | 1.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.9 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 0.9 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 0.5 | GO:0021767 | mammillary body development(GO:0021767) |
| 0.1 | 1.0 | GO:0014034 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.1 | 0.3 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 1.1 | GO:0042026 | protein refolding(GO:0042026) |
| 0.1 | 1.2 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 1.0 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.1 | 0.7 | GO:0021794 | thalamus development(GO:0021794) |
| 0.1 | 0.8 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.1 | 0.3 | GO:0071869 | response to monoamine(GO:0071867) response to catecholamine(GO:0071869) response to epinephrine(GO:0071871) |
| 0.1 | 1.0 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 0.6 | GO:0071156 | regulation of cell cycle arrest(GO:0071156) |
| 0.1 | 0.8 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.1 | 0.4 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.1 | 0.4 | GO:0003348 | cardiac endothelial cell differentiation(GO:0003348) endocardial cell differentiation(GO:0060956) |
| 0.1 | 0.4 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.1 | 1.3 | GO:0005979 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.1 | 0.4 | GO:1900118 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.2 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.1 | 1.0 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 1.1 | GO:0030168 | platelet activation(GO:0030168) |
| 0.1 | 0.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.7 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 1.1 | GO:0007094 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) negative regulation of mitotic metaphase/anaphase transition(GO:0045841) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) |
| 0.0 | 0.2 | GO:1904086 | regulation of epiboly involved in gastrulation with mouth forming second(GO:1904086) |
| 0.0 | 0.4 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 2.3 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.8 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 3.2 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.9 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.1 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.0 | 1.3 | GO:0002455 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 0.0 | 1.0 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 1.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.3 | GO:0090050 | positive regulation of blood vessel endothelial cell migration(GO:0043536) positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 1.1 | GO:0060872 | semicircular canal development(GO:0060872) |
| 0.0 | 1.3 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.2 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.3 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.9 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.5 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 1.6 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 1.6 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.3 | GO:0010885 | regulation of cholesterol storage(GO:0010885) |
| 0.0 | 0.8 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 1.2 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.5 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.2 | GO:0090594 | inflammatory response to wounding(GO:0090594) |
| 0.0 | 1.2 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.0 | 0.1 | GO:0042823 | pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.0 | 0.7 | GO:0032402 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 0.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.7 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.3 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 0.7 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.4 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.2 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 0.0 | 0.2 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.6 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.0 | 0.2 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.3 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.0 | 0.2 | GO:0043490 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.2 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.6 | GO:0042129 | regulation of T cell proliferation(GO:0042129) |
| 0.0 | 0.4 | GO:0008345 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.0 | 0.1 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.0 | 0.2 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.7 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.1 | GO:0090113 | regulation of COPII vesicle coating(GO:0003400) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.0 | 0.5 | GO:0048663 | neuron fate commitment(GO:0048663) |
| 0.0 | 0.1 | GO:0046958 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.0 | 0.3 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.0 | 0.6 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.1 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.2 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.6 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.0 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.1 | GO:0031268 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.0 | 0.5 | GO:0001841 | neural tube formation(GO:0001841) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.9 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.4 | 1.6 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.4 | 1.6 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.3 | 1.5 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.2 | 1.2 | GO:0034359 | mature chylomicron(GO:0034359) |
| 0.2 | 0.9 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 1.4 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 0.5 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 0.9 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 1.3 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.1 | 0.6 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.7 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.1 | 2.0 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 1.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 1.2 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 0.8 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 1.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 1.0 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.2 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.6 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.2 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.0 | 0.3 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 8.1 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 2.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.3 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 1.5 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.3 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.5 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.8 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.3 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.4 | GO:0044447 | axoneme part(GO:0044447) |
| 0.0 | 0.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.0 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.1 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.0 | 1.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.0 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.3 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.6 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 1.8 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 1.2 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 1.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.7 | GO:0016592 | mediator complex(GO:0016592) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.5 | 1.5 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.3 | 2.4 | GO:0070888 | E-box binding(GO:0070888) |
| 0.3 | 1.3 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.2 | 1.2 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.2 | 1.6 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.2 | 5.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.2 | 1.2 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.2 | 0.7 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 0.7 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.2 | 0.7 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.2 | 0.8 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.2 | 1.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.2 | 1.4 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.2 | 2.0 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.1 | 1.3 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.1 | 0.9 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.1 | 1.1 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 0.4 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.1 | 0.9 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 1.2 | GO:0070325 | low-density lipoprotein particle receptor binding(GO:0050750) lipoprotein particle receptor binding(GO:0070325) |
| 0.1 | 1.1 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 0.5 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.1 | 1.0 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.1 | 0.3 | GO:0000703 | oxidized pyrimidine nucleobase lesion DNA N-glycosylase activity(GO:0000703) |
| 0.1 | 0.3 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.1 | 1.7 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.1 | 0.6 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 1.3 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 0.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.1 | 1.0 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 0.3 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 1.1 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.5 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.1 | 1.4 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 0.6 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 0.3 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 0.4 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 0.6 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.4 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.1 | 0.2 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.1 | 1.6 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.9 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 1.0 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.7 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.7 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.6 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.4 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 1.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 4.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.0 | 0.2 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.7 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.4 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) |
| 0.0 | 0.8 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 4.2 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 2.1 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.3 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.3 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 6.9 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.7 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.2 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 1.5 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 1.5 | GO:0051536 | iron-sulfur cluster binding(GO:0051536) metal cluster binding(GO:0051540) |
| 0.0 | 0.5 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.3 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 1.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.4 | GO:0016411 | acylglycerol O-acyltransferase activity(GO:0016411) |
| 0.0 | 0.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 35.7 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 0.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 1.6 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.2 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.7 | GO:0016879 | ligase activity, forming carbon-nitrogen bonds(GO:0016879) |
| 0.0 | 0.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.9 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 2.6 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 2.0 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.3 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) phosphatidylinositol monophosphate phosphatase activity(GO:0052744) |
| 0.0 | 2.8 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.2 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.8 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 1.1 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 1.8 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 1.2 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 0.6 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 5.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 0.8 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 2.6 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.7 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.2 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.6 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 1.5 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.3 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 1.3 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.6 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.3 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.4 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.4 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.7 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.2 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.3 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.8 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.4 | 3.6 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.2 | 2.0 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 2.0 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 0.6 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 1.5 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 2.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 2.6 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 1.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 1.0 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 1.1 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 1.2 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 2.0 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.0 | 0.8 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 1.1 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 1.3 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.1 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.3 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.5 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.5 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.2 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |