Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
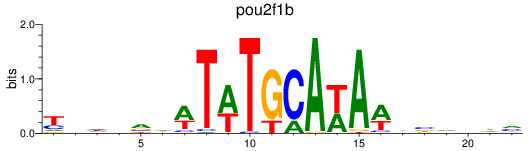
Results for pou2f1b
Z-value: 0.52
Transcription factors associated with pou2f1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pou2f1b
|
ENSDARG00000007996 | POU class 2 homeobox 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pou2f1b | dr11_v1_chr9_+_34350025_34350025 | 0.18 | 4.5e-01 | Click! |
Activity profile of pou2f1b motif
Sorted Z-values of pou2f1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_77506362 | 0.71 |
ENSDART00000174387
ENSDART00000181181 |
CABZ01087415.1
|
|
| chr20_+_34374735 | 0.70 |
ENSDART00000144090
|
si:dkeyp-11g8.3
|
si:dkeyp-11g8.3 |
| chr5_+_35458190 | 0.69 |
ENSDART00000051313
|
fbp1b
|
fructose-1,6-bisphosphatase 1b |
| chr23_+_22200467 | 0.64 |
ENSDART00000025414
|
slc2a1a
|
solute carrier family 2 (facilitated glucose transporter), member 1a |
| chr9_+_48755638 | 0.63 |
ENSDART00000047401
|
abcb11a
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11a |
| chr18_+_26422124 | 0.61 |
ENSDART00000060245
|
ctsh
|
cathepsin H |
| chr20_+_52492192 | 0.60 |
ENSDART00000057986
|
TSTA3 (1 of many)
|
zgc:100864 |
| chr18_+_22287084 | 0.58 |
ENSDART00000151919
ENSDART00000181644 |
ctcf
|
CCCTC-binding factor (zinc finger protein) |
| chr24_-_23758003 | 0.57 |
ENSDART00000178085
|
BX640462.2
|
Danio rerio minichromosome maintenance domain containing 2 (mcmdc2), mRNA. |
| chr16_+_52847079 | 0.57 |
ENSDART00000163151
|
cep72
|
centrosomal protein 72 |
| chr11_-_25418856 | 0.52 |
ENSDART00000013714
|
gata1a
|
GATA binding protein 1a |
| chr9_-_33062891 | 0.52 |
ENSDART00000161182
|
si:ch211-125e6.5
|
si:ch211-125e6.5 |
| chr22_+_29994093 | 0.51 |
ENSDART00000104778
|
si:dkey-286j15.3
|
si:dkey-286j15.3 |
| chr9_-_33063083 | 0.50 |
ENSDART00000048550
|
si:ch211-125e6.5
|
si:ch211-125e6.5 |
| chr19_-_1948236 | 0.49 |
ENSDART00000163344
|
znrf2a
|
zinc and ring finger 2a |
| chr8_-_50979047 | 0.45 |
ENSDART00000184788
ENSDART00000180906 |
zgc:91909
|
zgc:91909 |
| chr13_+_13930263 | 0.45 |
ENSDART00000079154
|
rpia
|
ribose 5-phosphate isomerase A (ribose 5-phosphate epimerase) |
| chr12_+_2676878 | 0.44 |
ENSDART00000185909
|
antxr1c
|
anthrax toxin receptor 1c |
| chr20_+_4221978 | 0.44 |
ENSDART00000171898
|
ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr5_+_42280372 | 0.44 |
ENSDART00000142855
|
tbx6l
|
T-box 6, like |
| chr15_-_43873005 | 0.44 |
ENSDART00000190326
|
NOX4
|
NADPH oxidase 4 |
| chr7_+_42935126 | 0.43 |
ENSDART00000157747
|
BX284696.1
|
|
| chr21_-_44512893 | 0.43 |
ENSDART00000166853
|
zgc:136410
|
zgc:136410 |
| chr25_+_22320738 | 0.42 |
ENSDART00000073566
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr8_-_44238526 | 0.42 |
ENSDART00000061115
ENSDART00000132473 ENSDART00000138019 ENSDART00000133021 |
piwil1
|
piwi-like RNA-mediated gene silencing 1 |
| chr24_-_12770357 | 0.42 |
ENSDART00000060826
|
ipo4
|
importin 4 |
| chr22_-_9906790 | 0.41 |
ENSDART00000105936
|
si:dkey-253d23.8
|
si:dkey-253d23.8 |
| chr25_-_18454016 | 0.38 |
ENSDART00000005877
|
cpa1
|
carboxypeptidase A1 (pancreatic) |
| chr16_-_17251898 | 0.38 |
ENSDART00000114272
|
nop2
|
NOP2 nucleolar protein homolog (yeast) |
| chr5_+_50879545 | 0.38 |
ENSDART00000128402
|
nol6
|
nucleolar protein 6 (RNA-associated) |
| chr5_+_35458346 | 0.38 |
ENSDART00000141239
|
erlin2
|
ER lipid raft associated 2 |
| chr21_-_23331619 | 0.37 |
ENSDART00000007806
|
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr12_+_22560067 | 0.37 |
ENSDART00000172066
|
polr2a
|
polymerase (RNA) II (DNA directed) polypeptide A |
| chr13_-_25198025 | 0.36 |
ENSDART00000159585
ENSDART00000144227 |
adka
|
adenosine kinase a |
| chr7_+_1579510 | 0.35 |
ENSDART00000190525
|
supt16h
|
SPT16 homolog, facilitates chromatin remodeling subunit |
| chr3_+_37112693 | 0.35 |
ENSDART00000055228
ENSDART00000144278 ENSDART00000138079 |
psmc3ip
|
PSMC3 interacting protein |
| chr10_+_38643304 | 0.35 |
ENSDART00000067447
|
mmp30
|
matrix metallopeptidase 30 |
| chr4_-_8043839 | 0.34 |
ENSDART00000190047
ENSDART00000057567 |
si:ch211-240l19.5
|
si:ch211-240l19.5 |
| chr10_-_41157135 | 0.33 |
ENSDART00000134851
|
aak1b
|
AP2 associated kinase 1b |
| chr5_+_11840905 | 0.33 |
ENSDART00000030444
|
tesca
|
tescalcin a |
| chr2_+_27394798 | 0.32 |
ENSDART00000115071
|
selenop2
|
selenoprotein P2 |
| chr6_-_55399214 | 0.32 |
ENSDART00000168367
|
ctsa
|
cathepsin A |
| chr5_-_11809710 | 0.32 |
ENSDART00000186998
ENSDART00000181363 ENSDART00000180681 |
nf2a
|
neurofibromin 2a (merlin) |
| chr5_-_68244564 | 0.32 |
ENSDART00000169350
|
CABZ01083944.1
|
|
| chr2_-_22993601 | 0.32 |
ENSDART00000125049
ENSDART00000099735 |
mllt1b
|
MLLT1, super elongation complex subunit b |
| chr7_-_6470431 | 0.31 |
ENSDART00000081359
|
zgc:110425
|
zgc:110425 |
| chr20_-_15099579 | 0.31 |
ENSDART00000127213
|
fmo5
|
flavin containing monooxygenase 5 |
| chr5_+_4806851 | 0.31 |
ENSDART00000067599
|
angptl2a
|
angiopoietin-like 2a |
| chr20_+_10538025 | 0.30 |
ENSDART00000129762
|
serpina1l
|
serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1, like |
| chr4_-_1801519 | 0.30 |
ENSDART00000188604
ENSDART00000135749 |
nudt4b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4b |
| chr4_+_23127284 | 0.30 |
ENSDART00000122675
|
mdm2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr25_-_9013963 | 0.30 |
ENSDART00000073912
|
rag2
|
recombination activating gene 2 |
| chr21_-_37733571 | 0.30 |
ENSDART00000176214
|
mpp1
|
membrane protein, palmitoylated 1 |
| chr20_-_14941046 | 0.30 |
ENSDART00000152775
ENSDART00000152516 ENSDART00000021394 |
mettl13
|
methyltransferase like 13 |
| chr2_-_4874966 | 0.29 |
ENSDART00000165296
|
tfr1a
|
transferrin receptor 1a |
| chr10_+_44824072 | 0.29 |
ENSDART00000162669
|
slc4a5
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 5 |
| chr3_+_49021079 | 0.29 |
ENSDART00000162012
|
zgc:163083
|
zgc:163083 |
| chr23_+_44665230 | 0.29 |
ENSDART00000144958
|
camta2
|
calmodulin binding transcription activator 2 |
| chr4_+_9592486 | 0.29 |
ENSDART00000080829
|
hspa14
|
heat shock protein 14 |
| chr4_-_64336270 | 0.29 |
ENSDART00000131672
|
znf1147
|
zinc finger protein 1147 |
| chr24_+_9178064 | 0.28 |
ENSDART00000142971
|
dlgap1b
|
discs, large (Drosophila) homolog-associated protein 1b |
| chr17_+_8542203 | 0.28 |
ENSDART00000158873
|
CU462878.2
|
|
| chr1_+_32054159 | 0.28 |
ENSDART00000181442
|
sts
|
steroid sulfatase (microsomal), isozyme S |
| chr10_-_40819022 | 0.28 |
ENSDART00000076316
ENSDART00000141329 |
zgc:113625
|
zgc:113625 |
| chr1_-_39859626 | 0.28 |
ENSDART00000053763
|
dctd
|
dCMP deaminase |
| chr21_+_36623162 | 0.28 |
ENSDART00000027459
|
grk6
|
G protein-coupled receptor kinase 6 |
| chr13_+_27768898 | 0.28 |
ENSDART00000029521
|
asrgl1
|
asparaginase like 1 |
| chr13_+_44857087 | 0.27 |
ENSDART00000017770
|
zbtb8os
|
zinc finger and BTB domain containing 8 opposite strand |
| chr3_+_49043917 | 0.27 |
ENSDART00000158212
|
zgc:92161
|
zgc:92161 |
| chr4_+_9178913 | 0.27 |
ENSDART00000168558
|
nfyba
|
nuclear transcription factor Y, beta a |
| chr23_+_39695827 | 0.27 |
ENSDART00000113893
ENSDART00000186679 |
tmco4
|
transmembrane and coiled-coil domains 4 |
| chr8_-_22965916 | 0.27 |
ENSDART00000143791
|
emilin3a
|
elastin microfibril interfacer 3a |
| chr12_+_34768106 | 0.26 |
ENSDART00000126420
ENSDART00000174433 |
slc38a10
|
solute carrier family 38, member 10 |
| chr22_+_14836291 | 0.26 |
ENSDART00000122740
|
gtpbp1l
|
GTP binding protein 1, like |
| chr21_+_30721733 | 0.26 |
ENSDART00000040443
|
zgc:110224
|
zgc:110224 |
| chr15_+_29393519 | 0.26 |
ENSDART00000193488
ENSDART00000112375 |
gdpd5b
|
glycerophosphodiester phosphodiesterase domain containing 5b |
| chr15_-_37867995 | 0.26 |
ENSDART00000192698
|
si:dkey-238d18.4
|
si:dkey-238d18.4 |
| chr4_+_51347000 | 0.26 |
ENSDART00000161859
|
si:dkeyp-82b4.3
|
si:dkeyp-82b4.3 |
| chr19_-_1947403 | 0.26 |
ENSDART00000113951
ENSDART00000151293 ENSDART00000134074 |
znrf2a
|
zinc and ring finger 2a |
| chr4_+_18707174 | 0.25 |
ENSDART00000142343
|
wnt7ba
|
wingless-type MMTV integration site family, member 7Ba |
| chr4_+_42604252 | 0.25 |
ENSDART00000184850
|
CR925768.1
|
|
| chr4_-_64141714 | 0.25 |
ENSDART00000128628
|
BX914205.3
|
|
| chr10_+_22918338 | 0.25 |
ENSDART00000167874
ENSDART00000171298 |
zgc:103508
|
zgc:103508 |
| chr4_+_25410293 | 0.25 |
ENSDART00000046253
ENSDART00000123680 |
prkcq
|
protein kinase C, theta |
| chr15_-_41700406 | 0.25 |
ENSDART00000181460
|
si:ch211-276c2.4
|
si:ch211-276c2.4 |
| chr19_-_25772980 | 0.24 |
ENSDART00000052393
|
pard6gb
|
par-6 family cell polarity regulator gamma b |
| chr12_-_22238004 | 0.24 |
ENSDART00000038310
|
ormdl3
|
ORMDL sphingolipid biosynthesis regulator 3 |
| chr1_-_56080112 | 0.24 |
ENSDART00000075469
ENSDART00000161473 |
c3a.6
|
complement component c3a, duplicate 6 |
| chr10_-_29831944 | 0.24 |
ENSDART00000063923
ENSDART00000136264 |
zpr1
|
ZPR1 zinc finger |
| chr2_-_55298075 | 0.24 |
ENSDART00000186404
ENSDART00000149062 |
rab8a
|
RAB8A, member RAS oncogene family |
| chr15_+_42995051 | 0.24 |
ENSDART00000183396
|
acsl3a
|
acyl-CoA synthetase long chain family member 3a |
| chr25_+_35019693 | 0.24 |
ENSDART00000046218
|
flnca
|
filamin C, gamma a (actin binding protein 280) |
| chr12_-_11593436 | 0.24 |
ENSDART00000138954
|
plekha1b
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1b |
| chr11_+_25596038 | 0.24 |
ENSDART00000140856
|
ccdc120
|
coiled-coil domain containing 120 |
| chr4_+_62679430 | 0.24 |
ENSDART00000145324
|
znf1131
|
zinc finger protein 1131 |
| chr17_+_23311377 | 0.24 |
ENSDART00000128073
|
ppp1r3ca
|
protein phosphatase 1, regulatory subunit 3Ca |
| chr16_+_38394371 | 0.24 |
ENSDART00000137954
|
cd83
|
CD83 molecule |
| chr19_-_27550768 | 0.23 |
ENSDART00000142313
|
si:dkeyp-46h3.8
|
si:dkeyp-46h3.8 |
| chr17_-_50020672 | 0.23 |
ENSDART00000188942
|
filip1a
|
filamin A interacting protein 1a |
| chr15_-_14083028 | 0.23 |
ENSDART00000147796
ENSDART00000043492 ENSDART00000133080 |
trappc6bl
|
trafficking protein particle complex 6b-like |
| chr4_-_26413391 | 0.23 |
ENSDART00000145955
|
b4galnt3a
|
beta-1,4-N-acetyl-galactosaminyl transferase 3a |
| chr24_+_23840821 | 0.23 |
ENSDART00000128595
ENSDART00000127188 |
pola1
|
polymerase (DNA directed), alpha 1 |
| chr18_-_15551360 | 0.23 |
ENSDART00000159915
ENSDART00000172690 |
ppfibp1b
|
PTPRF interacting protein, binding protein 1b (liprin beta 1) |
| chr2_-_43130812 | 0.23 |
ENSDART00000039079
ENSDART00000056162 |
ftr13
|
finTRIM family, member 13 |
| chr15_+_19838458 | 0.23 |
ENSDART00000101204
|
alcamb
|
activated leukocyte cell adhesion molecule b |
| chr15_+_28106498 | 0.23 |
ENSDART00000041707
|
unc119a
|
unc-119 homolog a (C. elegans) |
| chr16_+_4078240 | 0.23 |
ENSDART00000160890
|
inpp5b
|
inositol polyphosphate-5-phosphatase B |
| chr17_+_16046314 | 0.22 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr19_-_29294457 | 0.22 |
ENSDART00000130815
ENSDART00000103437 |
e2f3
|
E2F transcription factor 3 |
| chr11_-_29658396 | 0.22 |
ENSDART00000183947
|
rpl22
|
ribosomal protein L22 |
| chr6_-_38930726 | 0.22 |
ENSDART00000154151
|
hdac7b
|
histone deacetylase 7b |
| chr10_+_9595575 | 0.22 |
ENSDART00000091780
ENSDART00000184287 |
rc3h2
|
ring finger and CCCH-type domains 2 |
| chr25_+_13662606 | 0.22 |
ENSDART00000008989
|
ccdc113
|
coiled-coil domain containing 113 |
| chr9_+_12934536 | 0.22 |
ENSDART00000134484
|
si:dkey-230p4.1
|
si:dkey-230p4.1 |
| chr13_-_30006260 | 0.22 |
ENSDART00000049624
|
wnt8b
|
wingless-type MMTV integration site family, member 8b |
| chr21_+_15295685 | 0.22 |
ENSDART00000135603
|
BX901878.1
|
|
| chr23_-_27235403 | 0.21 |
ENSDART00000134418
|
si:dkey-157g16.6
|
si:dkey-157g16.6 |
| chr18_-_3166726 | 0.21 |
ENSDART00000165002
|
aqp11
|
aquaporin 11 |
| chr6_+_33931095 | 0.21 |
ENSDART00000191909
|
orc1
|
origin recognition complex, subunit 1 |
| chr10_+_40214877 | 0.21 |
ENSDART00000109689
|
gramd1ba
|
GRAM domain containing 1Ba |
| chr2_+_10766744 | 0.21 |
ENSDART00000015379
|
gfi1aa
|
growth factor independent 1A transcription repressor a |
| chr12_-_10381282 | 0.21 |
ENSDART00000152788
|
mki67
|
marker of proliferation Ki-67 |
| chr15_-_34001000 | 0.21 |
ENSDART00000180464
|
VWDE
|
si:dkey-30e9.7 |
| chr2_+_43204919 | 0.21 |
ENSDART00000160077
ENSDART00000018729 ENSDART00000129134 ENSDART00000056402 |
pard3ab
|
par-3 family cell polarity regulator alpha, b |
| chr5_+_31350927 | 0.21 |
ENSDART00000028237
|
camkk1a
|
calcium/calmodulin-dependent protein kinase kinase 1, alpha a |
| chr15_-_26923993 | 0.21 |
ENSDART00000132459
|
ccdc9
|
coiled-coil domain containing 9 |
| chr10_+_31809226 | 0.21 |
ENSDART00000087898
|
foxo1b
|
forkhead box O1 b |
| chr25_+_25516016 | 0.21 |
ENSDART00000188980
|
phrf1
|
PHD and ring finger domains 1 |
| chr16_-_39131666 | 0.20 |
ENSDART00000075517
|
gdf6a
|
growth differentiation factor 6a |
| chr4_-_8035520 | 0.20 |
ENSDART00000146146
|
si:ch211-240l19.7
|
si:ch211-240l19.7 |
| chr16_-_46587938 | 0.20 |
ENSDART00000181433
|
BX323793.1
|
|
| chr4_-_59152337 | 0.20 |
ENSDART00000156770
|
znf1149
|
zinc finger protein 1149 |
| chr6_-_19683406 | 0.20 |
ENSDART00000158041
|
cfap52
|
cilia and flagella associated protein 52 |
| chr13_+_18545819 | 0.20 |
ENSDART00000101859
ENSDART00000110197 ENSDART00000136064 |
zgc:154058
|
zgc:154058 |
| chr3_-_49138004 | 0.20 |
ENSDART00000167173
|
gipc1
|
GIPC PDZ domain containing family, member 1 |
| chr1_+_26110985 | 0.19 |
ENSDART00000054208
|
mtap
|
methylthioadenosine phosphorylase |
| chr7_+_1579236 | 0.19 |
ENSDART00000172830
|
supt16h
|
SPT16 homolog, facilitates chromatin remodeling subunit |
| chr8_+_43016714 | 0.19 |
ENSDART00000142671
|
rassf2a
|
Ras association (RalGDS/AF-6) domain family member 2a |
| chr11_-_21404044 | 0.19 |
ENSDART00000080116
|
ikbke
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
| chr3_+_7771420 | 0.19 |
ENSDART00000156809
ENSDART00000156309 |
hook2
|
hook microtubule-tethering protein 2 |
| chr5_+_29714786 | 0.19 |
ENSDART00000148314
|
ddx31
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 31 |
| chr5_+_28497956 | 0.19 |
ENSDART00000191935
|
nfr
|
notochord formation related |
| chr21_-_34926619 | 0.19 |
ENSDART00000065337
ENSDART00000192740 |
kif20a
|
kinesin family member 20A |
| chr15_-_14038227 | 0.19 |
ENSDART00000139068
|
zgc:114130
|
zgc:114130 |
| chr22_-_37834312 | 0.19 |
ENSDART00000076128
|
ppp1r2
|
protein phosphatase 1, regulatory (inhibitor) subunit 2 |
| chr23_+_12455295 | 0.19 |
ENSDART00000143752
ENSDART00000135785 |
si:ch211-153a8.4
|
si:ch211-153a8.4 |
| chr13_+_19283936 | 0.19 |
ENSDART00000111462
|
eif3s10
|
eukaryotic translation initiation factor 3, subunit 10 (theta) |
| chr18_+_38192499 | 0.18 |
ENSDART00000191849
|
nucb2b
|
nucleobindin 2b |
| chr25_+_16116740 | 0.18 |
ENSDART00000139778
|
far1
|
fatty acyl CoA reductase 1 |
| chr4_-_5332814 | 0.18 |
ENSDART00000144225
|
osgep
|
O-sialoglycoprotein endopeptidase |
| chr14_-_7409364 | 0.18 |
ENSDART00000036463
|
dnd1
|
DND microRNA-mediated repression inhibitor 1 |
| chr24_-_20641000 | 0.18 |
ENSDART00000166135
|
zbtb47b
|
zinc finger and BTB domain containing 47b |
| chr11_+_13024002 | 0.18 |
ENSDART00000104113
|
btf3l4
|
basic transcription factor 3-like 4 |
| chr12_+_3875901 | 0.18 |
ENSDART00000152514
|
nif3l1
|
NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae) |
| chr17_-_28101880 | 0.18 |
ENSDART00000149458
ENSDART00000149226 ENSDART00000085758 |
kdm1a
|
lysine (K)-specific demethylase 1a |
| chr15_-_23814330 | 0.18 |
ENSDART00000153843
|
si:ch211-167j9.5
|
si:ch211-167j9.5 |
| chr16_+_47428721 | 0.18 |
ENSDART00000180597
ENSDART00000032309 |
slc25a32b
|
solute carrier family 25 (mitochondrial folate carrier), member 32b |
| chr13_-_1130096 | 0.18 |
ENSDART00000010261
|
pno1
|
partner of NOB1 homolog |
| chr5_+_29715040 | 0.18 |
ENSDART00000192563
|
ddx31
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 31 |
| chr9_-_33608427 | 0.18 |
ENSDART00000100849
|
caska
|
calcium/calmodulin-dependent serine protein kinase a |
| chr5_+_29726428 | 0.18 |
ENSDART00000143183
|
ddx31
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 31 |
| chr1_+_39859782 | 0.18 |
ENSDART00000149984
|
irf2a
|
interferon regulatory factor 2a |
| chr4_+_42450386 | 0.17 |
ENSDART00000168211
|
si:dkey-11d20.1
|
si:dkey-11d20.1 |
| chr5_+_27583117 | 0.17 |
ENSDART00000180340
|
zmat4a
|
zinc finger, matrin-type 4a |
| chr4_-_49801268 | 0.17 |
ENSDART00000185457
|
si:ch211-197e7.1
|
si:ch211-197e7.1 |
| chr7_-_11384279 | 0.17 |
ENSDART00000102515
ENSDART00000172796 |
mesd
|
mesoderm development LRP chaperone |
| chr19_+_42432625 | 0.17 |
ENSDART00000076938
|
pogza
|
pogo transposable element derived with ZNF domain a |
| chr8_-_37391372 | 0.17 |
ENSDART00000190407
ENSDART00000009569 |
slc12a5b
|
solute carrier family 12 (potassium/chloride transporter), member 5b |
| chr20_-_13142357 | 0.17 |
ENSDART00000189220
|
ints7
|
integrator complex subunit 7 |
| chr11_-_21404358 | 0.17 |
ENSDART00000129062
|
ikbke
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
| chr4_+_35316044 | 0.17 |
ENSDART00000109741
|
BX000703.1
|
|
| chr24_+_32668675 | 0.17 |
ENSDART00000156638
ENSDART00000155973 |
si:ch211-282b22.1
|
si:ch211-282b22.1 |
| chr6_+_29410986 | 0.17 |
ENSDART00000065293
|
usp13
|
ubiquitin specific peptidase 13 |
| chr22_-_36530902 | 0.17 |
ENSDART00000056188
|
polr2h
|
info polymerase (RNA) II (DNA directed) polypeptide H |
| chr13_+_18507592 | 0.17 |
ENSDART00000142622
|
si:ch211-198a12.6
|
si:ch211-198a12.6 |
| chr2_+_20868286 | 0.17 |
ENSDART00000062591
|
odr4
|
odr-4 GPCR localization factor homolog |
| chr19_-_43659779 | 0.17 |
ENSDART00000144058
|
si:ch211-193k19.2
|
si:ch211-193k19.2 |
| chr5_+_38720140 | 0.17 |
ENSDART00000142103
|
si:dkey-58f10.7
|
si:dkey-58f10.7 |
| chr6_+_40922572 | 0.17 |
ENSDART00000133599
ENSDART00000002728 ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr2_-_32513538 | 0.17 |
ENSDART00000056640
|
abcf2a
|
ATP-binding cassette, sub-family F (GCN20), member 2a |
| chr14_-_21064199 | 0.17 |
ENSDART00000172099
|
si:dkey-74k8.3
|
si:dkey-74k8.3 |
| chr20_+_572037 | 0.16 |
ENSDART00000028062
ENSDART00000152736 ENSDART00000031759 ENSDART00000162198 |
smyd2b
|
SET and MYND domain containing 2b |
| chr24_+_38534550 | 0.16 |
ENSDART00000105677
|
zgc:154125
|
zgc:154125 |
| chr2_+_37838259 | 0.16 |
ENSDART00000136796
|
parp2
|
poly (ADP-ribose) polymerase 2 |
| chr5_-_1487256 | 0.16 |
ENSDART00000149599
ENSDART00000148411 ENSDART00000092087 ENSDART00000148464 |
golga2
|
golgin A2 |
| chr2_+_50862527 | 0.16 |
ENSDART00000169800
ENSDART00000158847 ENSDART00000160900 |
adcyap1r1a
|
adenylate cyclase activating polypeptide 1a (pituitary) receptor type I |
| chr12_+_34854562 | 0.16 |
ENSDART00000130366
|
si:dkey-21c1.4
|
si:dkey-21c1.4 |
| chr4_+_5341592 | 0.16 |
ENSDART00000123375
ENSDART00000067371 |
zgc:113263
|
zgc:113263 |
| chr8_+_26268726 | 0.16 |
ENSDART00000180883
|
slc26a6
|
solute carrier family 26, member 6 |
| chr3_-_33422738 | 0.16 |
ENSDART00000075493
|
ccdc103
|
coiled-coil domain containing 103 |
| chr5_+_27583445 | 0.16 |
ENSDART00000136488
|
zmat4a
|
zinc finger, matrin-type 4a |
| chr13_-_49802194 | 0.16 |
ENSDART00000148722
|
b3galnt2
|
beta-1,3-N-acetylgalactosaminyltransferase 2 |
| chr17_-_32743044 | 0.16 |
ENSDART00000135517
|
cenpf
|
centromere protein F |
| chr24_-_12958668 | 0.15 |
ENSDART00000178982
|
FITM1 (1 of many)
|
Danio rerio fat storage inducing transmembrane protein 1 (LOC792443), mRNA. |
| chr5_+_7564644 | 0.15 |
ENSDART00000192173
|
CABZ01039096.1
|
|
| chr24_+_9612218 | 0.15 |
ENSDART00000175671
|
nphp3
|
nephronophthisis 3 |
| chr19_-_40186328 | 0.15 |
ENSDART00000087474
|
efcab1
|
EF-hand calcium binding domain 1 |
| chr4_+_5832311 | 0.15 |
ENSDART00000121743
ENSDART00000158233 ENSDART00000165187 |
si:ch73-352p4.5
|
si:ch73-352p4.5 |
| chr1_-_44638058 | 0.15 |
ENSDART00000081835
|
slc43a1b
|
solute carrier family 43 (amino acid system L transporter), member 1b |
| chr3_+_24062748 | 0.15 |
ENSDART00000188574
|
cbx1a
|
chromobox homolog 1a (HP1 beta homolog Drosophila) |
Network of associatons between targets according to the STRING database.
First level regulatory network of pou2f1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0005985 | sucrose metabolic process(GO:0005985) sucrose biosynthetic process(GO:0005986) disaccharide biosynthetic process(GO:0046351) |
| 0.2 | 0.6 | GO:0015722 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.1 | 0.5 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.1 | 0.4 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 0.3 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.1 | 0.3 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.1 | 0.4 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.1 | 0.4 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.1 | 0.6 | GO:0046323 | glucose import(GO:0046323) |
| 0.1 | 0.2 | GO:0002468 | dendritic cell antigen processing and presentation(GO:0002468) |
| 0.1 | 0.5 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.1 | 0.3 | GO:0015682 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.1 | 0.4 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.1 | 0.2 | GO:0060898 | eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.1 | 0.4 | GO:0035093 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 | 0.3 | GO:0006530 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.1 | 0.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 0.2 | GO:0090155 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.1 | 0.3 | GO:1901906 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.3 | GO:1904036 | negative regulation of epithelial cell apoptotic process(GO:1904036) |
| 0.1 | 0.3 | GO:1904398 | positive regulation of neuromuscular junction development(GO:1904398) |
| 0.1 | 0.2 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.1 | 0.2 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.1 | 0.2 | GO:0071867 | response to monoamine(GO:0071867) response to catecholamine(GO:0071869) response to epinephrine(GO:0071871) |
| 0.1 | 0.3 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.1 | 0.2 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.1 | 0.3 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 0.2 | GO:0043703 | retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.0 | 0.4 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.2 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.1 | GO:0035971 | peptidyl-histidine dephosphorylation(GO:0035971) |
| 0.0 | 0.1 | GO:0060912 | cardiac cell fate specification(GO:0060912) |
| 0.0 | 0.2 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.1 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.0 | 0.4 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.2 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.1 | GO:0015824 | L-alanine transport(GO:0015808) proline transport(GO:0015824) |
| 0.0 | 0.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.5 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 | 0.1 | GO:0031441 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.2 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.6 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.2 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.6 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.2 | GO:0021885 | forebrain cell migration(GO:0021885) |
| 0.0 | 0.2 | GO:0060965 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.0 | 0.2 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.4 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.0 | 0.1 | GO:1901376 | mycotoxin metabolic process(GO:0043385) aflatoxin metabolic process(GO:0046222) organic heteropentacyclic compound metabolic process(GO:1901376) |
| 0.0 | 0.3 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.2 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.0 | 0.2 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.2 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.2 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.1 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.3 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.3 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.1 | GO:1900182 | positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.0 | 0.3 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 0.1 | GO:0097638 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.1 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.2 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.3 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.0 | 0.2 | GO:0008105 | asymmetric protein localization(GO:0008105) apical protein localization(GO:0045176) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.1 | GO:0006844 | acyl carnitine transport(GO:0006844) |
| 0.0 | 0.1 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 0.3 | GO:0060969 | nucleosome positioning(GO:0016584) negative regulation of chromatin silencing(GO:0031936) negative regulation of gene silencing(GO:0060969) |
| 0.0 | 0.3 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 0.1 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0006867 | asparagine transport(GO:0006867) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:0042269 | natural killer cell mediated immunity(GO:0002228) regulation of natural killer cell mediated immunity(GO:0002715) positive regulation of natural killer cell mediated immunity(GO:0002717) natural killer cell mediated cytotoxicity(GO:0042267) regulation of natural killer cell mediated cytotoxicity(GO:0042269) positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.0 | 0.2 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.2 | GO:1904356 | regulation of telomere maintenance via telomere lengthening(GO:1904356) |
| 0.0 | 0.1 | GO:0090113 | regulation of COPII vesicle coating(GO:0003400) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.0 | 0.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.0 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.1 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.0 | 0.1 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.2 | GO:0071594 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.0 | 0.1 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.4 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.0 | 0.1 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.0 | 0.1 | GO:0003422 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.0 | 0.3 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.0 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) positive regulation of macrophage chemotaxis(GO:0010759) skeletal muscle cell proliferation(GO:0014856) regulation of skeletal muscle cell proliferation(GO:0014857) mononuclear cell migration(GO:0071674) regulation of mononuclear cell migration(GO:0071675) positive regulation of lamellipodium organization(GO:1902745) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0034456 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.1 | 0.3 | GO:0097519 | DNA recombinase complex(GO:0097519) |
| 0.1 | 0.2 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.1 | 0.2 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.5 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.5 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.2 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.0 | 0.2 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.0 | 0.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.0 | 0.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.2 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.1 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.1 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.2 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.1 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.2 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.4 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.1 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.0 | 0.3 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.2 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.2 | GO:0019814 | immunoglobulin complex(GO:0019814) |
| 0.0 | 0.0 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.2 | 0.6 | GO:0015126 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.1 | 0.4 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.1 | 0.4 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.1 | 0.3 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 0.5 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.1 | 0.3 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.1 | 0.3 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 0.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.2 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.1 | 0.3 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 0.2 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.1 | 0.3 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.1 | 0.4 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.6 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) monosaccharide transmembrane transporter activity(GO:0015145) hexose transmembrane transporter activity(GO:0015149) |
| 0.0 | 0.1 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.1 | GO:0101006 | protein histidine phosphatase activity(GO:0101006) |
| 0.0 | 0.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 0.2 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.0 | 0.4 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.2 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.2 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.2 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.2 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.1 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.1 | GO:0015193 | L-alanine transmembrane transporter activity(GO:0015180) L-proline transmembrane transporter activity(GO:0015193) alanine transmembrane transporter activity(GO:0022858) |
| 0.0 | 0.2 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.3 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.2 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.0 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.1 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.1 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.0 | 0.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.1 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.0 | 0.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.4 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.3 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.5 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.0 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.2 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.1 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.0 | 0.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.4 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.2 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.1 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.0 | 0.2 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.0 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0055102 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.0 | 0.1 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.4 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.3 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.2 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.4 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.4 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.4 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.3 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.4 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.3 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 1.5 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.2 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.7 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.1 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.3 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.8 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |