Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
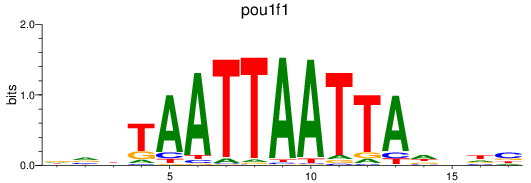
Results for pou1f1
Z-value: 0.34
Transcription factors associated with pou1f1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pou1f1
|
ENSDARG00000058924 | POU class 1 homeobox 1 |
|
pou1f1
|
ENSDARG00000110816 | POU class 1 homeobox 1 |
|
pou1f1
|
ENSDARG00000115109 | POU class 1 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pou1f1 | dr11_v1_chr9_-_19161982_19161982 | 0.12 | 6.3e-01 | Click! |
Activity profile of pou1f1 motif
Sorted Z-values of pou1f1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_22099536 | 2.86 |
ENSDART00000101923
|
CR391987.1
|
|
| chr12_-_17712393 | 2.01 |
ENSDART00000143534
ENSDART00000010144 |
pvalb2
|
parvalbumin 2 |
| chr9_-_22213297 | 1.67 |
ENSDART00000110656
ENSDART00000133149 |
crygm2d20
|
crystallin, gamma M2d20 |
| chr14_+_46313396 | 1.05 |
ENSDART00000047525
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr21_-_20939488 | 1.03 |
ENSDART00000039043
|
rgs7bpb
|
regulator of G protein signaling 7 binding protein b |
| chr25_+_31227747 | 0.86 |
ENSDART00000033872
|
tnni2a.1
|
troponin I type 2a (skeletal, fast), tandem duplicate 1 |
| chr5_-_44829719 | 0.81 |
ENSDART00000019104
|
fbp2
|
fructose-1,6-bisphosphatase 2 |
| chr2_+_2223837 | 0.79 |
ENSDART00000101038
ENSDART00000129354 |
tmie
|
transmembrane inner ear |
| chr9_+_18716485 | 0.77 |
ENSDART00000135125
|
serp2
|
stress-associated endoplasmic reticulum protein family member 2 |
| chr4_+_9669717 | 0.77 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr20_-_40755614 | 0.64 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr16_-_28856112 | 0.64 |
ENSDART00000078543
|
syt11b
|
synaptotagmin XIb |
| chr16_-_12173554 | 0.59 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr9_+_54290896 | 0.57 |
ENSDART00000149175
|
pou4f3
|
POU class 4 homeobox 3 |
| chr23_+_31405497 | 0.55 |
ENSDART00000053546
|
sh3bgrl2
|
SH3 domain binding glutamate-rich protein like 2 |
| chr4_-_5302866 | 0.52 |
ENSDART00000138590
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr9_+_34641237 | 0.50 |
ENSDART00000133996
|
shox
|
short stature homeobox |
| chr20_-_5291012 | 0.50 |
ENSDART00000122892
|
cyp46a1.3
|
cytochrome P450, family 46, subfamily A, polypeptide 1, tandem duplicate 3 |
| chr9_+_2574122 | 0.50 |
ENSDART00000166326
ENSDART00000191822 |
CIR1
|
si:ch73-167c12.2 |
| chr13_-_29420885 | 0.49 |
ENSDART00000024225
|
chata
|
choline O-acetyltransferase a |
| chr23_-_20051369 | 0.49 |
ENSDART00000049836
|
bgnb
|
biglycan b |
| chr5_-_41494831 | 0.49 |
ENSDART00000051081
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr7_+_24573721 | 0.47 |
ENSDART00000173938
ENSDART00000173681 |
si:dkeyp-75h12.7
|
si:dkeyp-75h12.7 |
| chr6_+_39905021 | 0.47 |
ENSDART00000064904
|
endou
|
endonuclease, polyU-specific |
| chr5_-_71705191 | 0.47 |
ENSDART00000187767
|
ak1
|
adenylate kinase 1 |
| chr16_+_46111849 | 0.47 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr19_+_10339538 | 0.47 |
ENSDART00000151808
ENSDART00000151235 |
rcvrn3
|
recoverin 3 |
| chr8_-_33114202 | 0.44 |
ENSDART00000098840
|
ralgps1
|
Ral GEF with PH domain and SH3 binding motif 1 |
| chr3_+_23092762 | 0.43 |
ENSDART00000142884
ENSDART00000024136 |
gngt2a
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2a |
| chr19_-_26869103 | 0.43 |
ENSDART00000089699
|
prrt1
|
proline-rich transmembrane protein 1 |
| chr18_+_924949 | 0.43 |
ENSDART00000170888
ENSDART00000193163 |
pkma
|
pyruvate kinase M1/2a |
| chr7_-_66868543 | 0.43 |
ENSDART00000149680
|
ampd3a
|
adenosine monophosphate deaminase 3a |
| chr11_+_30663300 | 0.40 |
ENSDART00000161662
|
ttbk1a
|
tau tubulin kinase 1a |
| chr1_-_22757145 | 0.40 |
ENSDART00000134719
|
prom1b
|
prominin 1 b |
| chr15_-_46779934 | 0.40 |
ENSDART00000085136
|
clcn2c
|
chloride channel 2c |
| chr6_-_11768198 | 0.40 |
ENSDART00000183463
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr18_+_39487486 | 0.39 |
ENSDART00000126978
|
acadl
|
acyl-CoA dehydrogenase long chain |
| chr5_+_2815021 | 0.39 |
ENSDART00000020472
|
hpda
|
4-hydroxyphenylpyruvate dioxygenase a |
| chr7_+_73397283 | 0.39 |
ENSDART00000174390
|
CABZ01081780.1
|
|
| chr6_-_28980756 | 0.38 |
ENSDART00000014661
|
glmnb
|
glomulin, FKBP associated protein b |
| chr16_-_22781446 | 0.37 |
ENSDART00000144107
|
lenep
|
lens epithelial protein |
| chr3_-_48716422 | 0.36 |
ENSDART00000164979
|
si:ch211-114m9.1
|
si:ch211-114m9.1 |
| chr21_-_22635245 | 0.36 |
ENSDART00000115224
ENSDART00000101782 |
nectin1a
|
nectin cell adhesion molecule 1a |
| chr13_+_24279021 | 0.36 |
ENSDART00000058629
|
acta1b
|
actin, alpha 1b, skeletal muscle |
| chr16_-_28658341 | 0.35 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr17_-_12336987 | 0.34 |
ENSDART00000172001
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr20_-_44557037 | 0.33 |
ENSDART00000140995
|
mfsd2b
|
major facilitator superfamily domain containing 2B |
| chr11_+_30057762 | 0.33 |
ENSDART00000164139
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr2_-_7666021 | 0.33 |
ENSDART00000180007
|
CABZ01021592.1
|
|
| chr1_-_22512063 | 0.32 |
ENSDART00000031546
ENSDART00000190987 |
chrna6
|
cholinergic receptor, nicotinic, alpha 6 |
| chr25_-_4146947 | 0.32 |
ENSDART00000129268
|
fads2
|
fatty acid desaturase 2 |
| chr1_-_15797663 | 0.32 |
ENSDART00000177122
|
SGCZ
|
sarcoglycan zeta |
| chr25_+_14087045 | 0.32 |
ENSDART00000155770
|
actc1c
|
actin, alpha, cardiac muscle 1c |
| chr22_-_13851297 | 0.31 |
ENSDART00000080306
|
s100b
|
S100 calcium binding protein, beta (neural) |
| chr20_-_34750363 | 0.31 |
ENSDART00000152845
|
znf395b
|
zinc finger protein 395b |
| chr11_-_29737088 | 0.30 |
ENSDART00000159828
|
si:ch211-218g23.6
|
si:ch211-218g23.6 |
| chr8_+_7359294 | 0.30 |
ENSDART00000121708
|
pcsk1nl
|
proprotein convertase subtilisin/kexin type 1 inhibitor, like |
| chr16_+_9540033 | 0.30 |
ENSDART00000149574
|
ca14
|
carbonic anhydrase XIV |
| chr4_+_22480169 | 0.30 |
ENSDART00000146272
ENSDART00000066904 |
ndufb2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2 |
| chr19_+_43297546 | 0.30 |
ENSDART00000168002
|
laptm5
|
lysosomal protein transmembrane 5 |
| chr16_-_38118003 | 0.29 |
ENSDART00000058667
|
si:dkey-23o4.6
|
si:dkey-23o4.6 |
| chr9_-_42418470 | 0.29 |
ENSDART00000144353
|
calcrla
|
calcitonin receptor-like a |
| chr16_+_36671064 | 0.29 |
ENSDART00000109703
|
col6a4a
|
collagen, type VI, alpha 4a |
| chr5_-_30382925 | 0.28 |
ENSDART00000125381
|
gig2o
|
grass carp reovirus (GCRV)-induced gene 2o |
| chr3_-_20040636 | 0.28 |
ENSDART00000104118
|
atxn7l3
|
ataxin 7-like 3 |
| chr5_+_69747417 | 0.27 |
ENSDART00000153717
|
si:ch211-275j6.5
|
si:ch211-275j6.5 |
| chr1_+_44127292 | 0.27 |
ENSDART00000160542
|
cabp2a
|
calcium binding protein 2a |
| chr6_+_52350443 | 0.27 |
ENSDART00000151612
ENSDART00000151349 |
si:ch211-239j9.1
|
si:ch211-239j9.1 |
| chr10_+_26747755 | 0.27 |
ENSDART00000100329
|
f9b
|
coagulation factor IXb |
| chr16_-_12173399 | 0.27 |
ENSDART00000142574
|
clstn3
|
calsyntenin 3 |
| chr2_-_9059955 | 0.26 |
ENSDART00000022768
|
ak5
|
adenylate kinase 5 |
| chr7_+_23515966 | 0.26 |
ENSDART00000186893
ENSDART00000186189 |
zgc:109889
|
zgc:109889 |
| chr3_+_23029934 | 0.26 |
ENSDART00000110343
|
nags
|
N-acetylglutamate synthase |
| chr9_-_43538328 | 0.26 |
ENSDART00000140526
|
znf385b
|
zinc finger protein 385B |
| chr1_-_23040358 | 0.25 |
ENSDART00000146871
|
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr12_-_558201 | 0.25 |
ENSDART00000168586
ENSDART00000158355 |
bsk146
|
brain specific kinase 146 |
| chr17_+_16564921 | 0.25 |
ENSDART00000151904
|
foxn3
|
forkhead box N3 |
| chr21_-_43015383 | 0.25 |
ENSDART00000065097
|
dpysl3
|
dihydropyrimidinase-like 3 |
| chr13_+_9432501 | 0.25 |
ENSDART00000058064
|
zgc:123321
|
zgc:123321 |
| chr20_-_34750045 | 0.25 |
ENSDART00000186130
|
znf395b
|
zinc finger protein 395b |
| chr20_+_4060839 | 0.25 |
ENSDART00000178565
|
TRIM67
|
tripartite motif containing 67 |
| chr12_+_3022882 | 0.24 |
ENSDART00000122905
|
rac3b
|
Rac family small GTPase 3b |
| chr2_+_20430366 | 0.24 |
ENSDART00000155108
|
si:ch211-153l6.6
|
si:ch211-153l6.6 |
| chr10_-_36927594 | 0.24 |
ENSDART00000162046
|
sez6a
|
seizure related 6 homolog a |
| chr15_+_25438714 | 0.24 |
ENSDART00000154164
|
aifm4
|
apoptosis-inducing factor, mitochondrion-associated, 4 |
| chr21_-_14811058 | 0.23 |
ENSDART00000143100
|
phpt1
|
phosphohistidine phosphatase 1 |
| chr20_+_19512727 | 0.23 |
ENSDART00000063696
|
atraid
|
all-trans retinoic acid-induced differentiation factor |
| chr6_+_40714811 | 0.23 |
ENSDART00000153868
|
ccdc36
|
coiled-coil domain containing 36 |
| chr6_-_35779348 | 0.23 |
ENSDART00000191159
|
brinp3a.1
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3a, tandem duplicate 1 |
| chr1_+_25801648 | 0.23 |
ENSDART00000129471
|
gucy1b1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr12_-_41684729 | 0.23 |
ENSDART00000184461
|
jakmip3
|
Janus kinase and microtubule interacting protein 3 |
| chr1_+_39482588 | 0.23 |
ENSDART00000181790
|
tenm3
|
teneurin transmembrane protein 3 |
| chr1_+_51721851 | 0.23 |
ENSDART00000040397
|
prdx2
|
peroxiredoxin 2 |
| chr5_-_6377865 | 0.23 |
ENSDART00000031775
|
zgc:73226
|
zgc:73226 |
| chr2_+_44571200 | 0.23 |
ENSDART00000098132
|
klhl24a
|
kelch-like family member 24a |
| chr20_+_32552912 | 0.22 |
ENSDART00000009691
|
scml4
|
Scm polycomb group protein like 4 |
| chr10_-_33621739 | 0.21 |
ENSDART00000142655
ENSDART00000128049 |
hunk
|
hormonally up-regulated Neu-associated kinase |
| chr24_+_40529346 | 0.21 |
ENSDART00000168548
|
CU929259.1
|
|
| chr5_-_55933420 | 0.21 |
ENSDART00000050966
|
slc25a46
|
solute carrier family 25, member 46 |
| chr12_+_1139690 | 0.21 |
ENSDART00000160442
|
CABZ01072885.1
|
|
| chr7_-_11596450 | 0.21 |
ENSDART00000173863
|
stard5
|
StAR-related lipid transfer (START) domain containing 5 |
| chr13_-_2010191 | 0.21 |
ENSDART00000161021
ENSDART00000124134 |
gfral
|
GDNF family receptor alpha like |
| chr3_+_26145013 | 0.21 |
ENSDART00000162546
ENSDART00000129561 |
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr9_+_54039006 | 0.21 |
ENSDART00000112441
|
tlr7
|
toll-like receptor 7 |
| chr24_-_26328721 | 0.20 |
ENSDART00000125468
|
apodb
|
apolipoprotein Db |
| chr13_-_29421331 | 0.20 |
ENSDART00000150228
|
chata
|
choline O-acetyltransferase a |
| chr16_-_31622777 | 0.20 |
ENSDART00000137311
ENSDART00000002930 |
phf20l1
|
PHD finger protein 20 like 1 |
| chr11_-_1935883 | 0.20 |
ENSDART00000172885
|
faim2b
|
Fas apoptotic inhibitory molecule 2b |
| chr21_+_22840246 | 0.20 |
ENSDART00000151621
|
birc2
|
baculoviral IAP repeat containing 2 |
| chr16_+_51326237 | 0.20 |
ENSDART00000168525
|
CU469384.1
|
|
| chr11_-_2838699 | 0.20 |
ENSDART00000066189
|
lhfpl5a
|
LHFPL tetraspan subfamily member 5a |
| chr8_-_2434282 | 0.20 |
ENSDART00000137262
ENSDART00000134044 |
vdac3
|
voltage-dependent anion channel 3 |
| chr5_-_2282256 | 0.19 |
ENSDART00000064012
|
ca4a
|
carbonic anhydrase IV a |
| chr5_-_42904329 | 0.19 |
ENSDART00000112807
|
cxcl20
|
chemokine (C-X-C motif) ligand 20 |
| chr22_+_1556948 | 0.19 |
ENSDART00000159050
|
si:ch211-255f4.8
|
si:ch211-255f4.8 |
| chr8_-_39952727 | 0.19 |
ENSDART00000181310
|
cabp1a
|
calcium binding protein 1a |
| chr8_-_25422186 | 0.18 |
ENSDART00000113492
ENSDART00000131736 |
kcnq2a
|
potassium voltage-gated channel, KQT-like subfamily, member 2a |
| chr17_+_45602640 | 0.18 |
ENSDART00000153822
|
si:ch211-202f3.4
|
si:ch211-202f3.4 |
| chr9_+_21826724 | 0.18 |
ENSDART00000101985
|
zgc:162944
|
zgc:162944 |
| chr6_+_612594 | 0.18 |
ENSDART00000150903
|
kynu
|
kynureninase |
| chr6_-_24053404 | 0.18 |
ENSDART00000168511
|
si:dkey-44g17.6
|
si:dkey-44g17.6 |
| chr8_+_8845932 | 0.18 |
ENSDART00000112028
|
si:ch211-180f4.1
|
si:ch211-180f4.1 |
| chr1_-_55118745 | 0.17 |
ENSDART00000133915
|
sertad2a
|
SERTA domain containing 2a |
| chr22_+_18469004 | 0.17 |
ENSDART00000061430
|
cilp2
|
cartilage intermediate layer protein 2 |
| chr15_+_9327252 | 0.17 |
ENSDART00000144381
|
sgcg
|
sarcoglycan, gamma |
| chr22_-_26865181 | 0.17 |
ENSDART00000138311
|
hmox2a
|
heme oxygenase 2a |
| chr14_-_413273 | 0.17 |
ENSDART00000163976
ENSDART00000179907 |
FAT4
|
FAT atypical cadherin 4 |
| chr4_+_5255041 | 0.17 |
ENSDART00000137966
|
ccdc167
|
coiled-coil domain containing 167 |
| chr14_-_7207961 | 0.17 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr3_+_36424055 | 0.17 |
ENSDART00000170318
|
si:ch1073-443f11.2
|
si:ch1073-443f11.2 |
| chr10_-_26766780 | 0.17 |
ENSDART00000146666
|
mcf2b
|
MCF.2 cell line derived transforming sequence b |
| chr2_+_16160906 | 0.17 |
ENSDART00000135783
|
selenoj
|
selenoprotein J |
| chr1_-_54063520 | 0.17 |
ENSDART00000171722
|
smdt1b
|
single-pass membrane protein with aspartate-rich tail 1b |
| chr11_+_40812590 | 0.16 |
ENSDART00000186690
|
errfi1a
|
ERBB receptor feedback inhibitor 1a |
| chr10_-_35236949 | 0.16 |
ENSDART00000145804
|
ypel2a
|
yippee-like 2a |
| chr2_-_1548330 | 0.16 |
ENSDART00000082155
ENSDART00000108481 ENSDART00000111272 |
dab1b
|
Dab, reelin signal transducer, homolog 1b (Drosophila) |
| chr20_+_41756996 | 0.16 |
ENSDART00000186393
|
fam184a
|
family with sequence similarity 184, member A |
| chr4_+_19700308 | 0.16 |
ENSDART00000027919
|
pax4
|
paired box 4 |
| chr9_+_34127005 | 0.16 |
ENSDART00000167384
ENSDART00000078065 |
f5
|
coagulation factor V |
| chr20_+_41664350 | 0.15 |
ENSDART00000186247
|
fam184a
|
family with sequence similarity 184, member A |
| chr18_+_35128685 | 0.15 |
ENSDART00000151579
|
si:ch211-195m9.3
|
si:ch211-195m9.3 |
| chr17_-_30702411 | 0.15 |
ENSDART00000114358
|
zgc:194392
|
zgc:194392 |
| chr22_-_31060579 | 0.15 |
ENSDART00000182376
|
cand2
|
cullin-associated and neddylation-dissociated 2 (putative) |
| chr8_+_26141680 | 0.15 |
ENSDART00000078334
|
celsr3
|
cadherin, EGF LAG seven-pass G-type receptor 3 |
| chr20_+_66857 | 0.15 |
ENSDART00000114999
|
lrfn2b
|
leucine rich repeat and fibronectin type III domain containing 2b |
| chr8_-_17987547 | 0.15 |
ENSDART00000112699
ENSDART00000061747 |
fpgt
|
fucose-1-phosphate guanylyltransferase |
| chr14_-_2209742 | 0.15 |
ENSDART00000054889
|
pcdh2ab5
|
protocadherin 2 alpha b 5 |
| chr21_+_21195487 | 0.14 |
ENSDART00000181746
ENSDART00000184832 |
rictorb
|
RPTOR independent companion of MTOR, complex 2b |
| chr11_-_26832685 | 0.14 |
ENSDART00000153519
|
iqsec1b
|
IQ motif and Sec7 domain 1b |
| chr16_+_12836143 | 0.14 |
ENSDART00000067741
|
cacng6b
|
calcium channel, voltage-dependent, gamma subunit 6b |
| chr15_+_34592215 | 0.14 |
ENSDART00000099776
|
tspan13a
|
tetraspanin 13a |
| chr1_-_58975098 | 0.14 |
ENSDART00000189899
|
CABZ01114581.1
|
|
| chr10_+_17776981 | 0.14 |
ENSDART00000141693
|
ccl19b
|
chemokine (C-C motif) ligand 19b |
| chr23_-_3511630 | 0.14 |
ENSDART00000019667
|
rnf114
|
ring finger protein 114 |
| chr18_-_47662696 | 0.14 |
ENSDART00000184260
|
CABZ01073963.1
|
|
| chr8_+_52637507 | 0.14 |
ENSDART00000163830
|
si:dkey-90l8.3
|
si:dkey-90l8.3 |
| chr13_+_7292061 | 0.13 |
ENSDART00000179504
|
CABZ01072077.1
|
Danio rerio neuroblast differentiation-associated protein AHNAK-like (LOC795051), mRNA. |
| chr21_+_45841731 | 0.13 |
ENSDART00000038657
|
faxdc2
|
fatty acid hydroxylase domain containing 2 |
| chr2_+_36007449 | 0.13 |
ENSDART00000161837
|
lamc2
|
laminin, gamma 2 |
| chr14_-_4145594 | 0.13 |
ENSDART00000077348
|
casp3b
|
caspase 3, apoptosis-related cysteine peptidase b |
| chr16_+_3068080 | 0.13 |
ENSDART00000160092
|
cyb5r4
|
cytochrome b5 reductase 4 |
| chr14_-_30141162 | 0.13 |
ENSDART00000053916
|
mtnr1ab
|
melatonin receptor 1A b |
| chr12_-_9132682 | 0.13 |
ENSDART00000066471
|
adam8b
|
ADAM metallopeptidase domain 8b |
| chr23_+_7710447 | 0.13 |
ENSDART00000168199
|
kif3b
|
kinesin family member 3B |
| chr4_-_2219705 | 0.13 |
ENSDART00000131046
|
si:ch73-278m9.1
|
si:ch73-278m9.1 |
| chr13_+_15656042 | 0.13 |
ENSDART00000134240
|
mark3a
|
MAP/microtubule affinity-regulating kinase 3a |
| chr22_+_23359369 | 0.13 |
ENSDART00000170886
|
dennd1b
|
DENN/MADD domain containing 1B |
| chr15_-_38154616 | 0.13 |
ENSDART00000099392
|
irgq2
|
immunity-related GTPase family, q2 |
| chr12_+_4222854 | 0.13 |
ENSDART00000144881
|
mapk7
|
mitogen-activated protein kinase 7 |
| chr21_-_32060993 | 0.12 |
ENSDART00000131651
|
si:ch211-160j14.2
|
si:ch211-160j14.2 |
| chr18_+_15616167 | 0.12 |
ENSDART00000080454
|
slc17a8
|
solute carrier family 17 (vesicular glutamate transporter), member 8 |
| chr25_-_19420949 | 0.12 |
ENSDART00000181338
|
map1ab
|
microtubule-associated protein 1Ab |
| chr2_-_7843451 | 0.12 |
ENSDART00000163265
|
si:ch211-38m6.7
|
si:ch211-38m6.7 |
| chr9_+_7724152 | 0.12 |
ENSDART00000061716
|
mnx2a
|
motor neuron and pancreas homeobox 2a |
| chr21_-_38853737 | 0.12 |
ENSDART00000184100
|
tlr22
|
toll-like receptor 22 |
| chr10_-_34916208 | 0.12 |
ENSDART00000187371
|
ccna1
|
cyclin A1 |
| chr1_+_6862917 | 0.12 |
ENSDART00000182953
|
erbb4a
|
erb-b2 receptor tyrosine kinase 4a |
| chr10_-_14556978 | 0.12 |
ENSDART00000126643
|
zgc:153395
|
zgc:153395 |
| chr9_-_40765868 | 0.12 |
ENSDART00000138634
|
abca12
|
ATP-binding cassette, sub-family A (ABC1), member 12 |
| chr7_-_22632690 | 0.12 |
ENSDART00000165245
|
si:dkey-112a7.4
|
si:dkey-112a7.4 |
| chr2_+_29491314 | 0.12 |
ENSDART00000181774
|
dlgap1a
|
discs, large (Drosophila) homolog-associated protein 1a |
| chr4_+_20051478 | 0.12 |
ENSDART00000143642
|
lamtor4
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 4 |
| chr7_+_49800849 | 0.11 |
ENSDART00000164838
ENSDART00000187888 |
PAMR1 (1 of many)
|
si:ch211-102l7.3 |
| chr23_-_41965557 | 0.11 |
ENSDART00000144183
|
slc1a7b
|
solute carrier family 1 (glutamate transporter), member 7b |
| chr22_+_1586060 | 0.11 |
ENSDART00000160793
|
si:ch211-255f4.11
|
si:ch211-255f4.11 |
| chr3_+_7808459 | 0.11 |
ENSDART00000162374
|
hook2
|
hook microtubule-tethering protein 2 |
| chr4_-_27897160 | 0.11 |
ENSDART00000066924
ENSDART00000066925 ENSDART00000193020 |
tbc1d22a
|
TBC1 domain family, member 22a |
| chr4_-_73136420 | 0.11 |
ENSDART00000160907
|
si:ch73-170d6.3
|
si:ch73-170d6.3 |
| chr7_-_71829649 | 0.11 |
ENSDART00000160449
|
cacnb2a
|
calcium channel, voltage-dependent, beta 2a |
| chr22_-_10121880 | 0.11 |
ENSDART00000002348
|
rdh5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr8_+_17987778 | 0.11 |
ENSDART00000133429
|
lrriq3
|
leucine-rich repeats and IQ motif containing 3 |
| chr19_+_12915498 | 0.11 |
ENSDART00000132892
|
cthrc1a
|
collagen triple helix repeat containing 1a |
| chr20_-_29864390 | 0.11 |
ENSDART00000161834
ENSDART00000132278 |
rnf144ab
|
ring finger protein 144ab |
| chr2_-_16216568 | 0.11 |
ENSDART00000173758
|
arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr9_+_45227028 | 0.11 |
ENSDART00000185579
|
adarb1b
|
adenosine deaminase, RNA-specific, B1b |
| chr9_-_25989989 | 0.11 |
ENSDART00000090052
|
arhgap15
|
Rho GTPase activating protein 15 |
| chr14_+_36521005 | 0.11 |
ENSDART00000192286
|
TENM3
|
si:dkey-237h12.3 |
| chr8_+_17987215 | 0.11 |
ENSDART00000113605
|
lrriq3
|
leucine-rich repeats and IQ motif containing 3 |
| chr2_+_20605925 | 0.11 |
ENSDART00000191510
|
olfml2bb
|
olfactomedin-like 2Bb |
| chr14_-_28534563 | 0.11 |
ENSDART00000054088
|
zgc:113364
|
zgc:113364 |
Network of associatons between targets according to the STRING database.
First level regulatory network of pou1f1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0005986 | sucrose metabolic process(GO:0005985) sucrose biosynthetic process(GO:0005986) disaccharide biosynthetic process(GO:0046351) |
| 0.2 | 0.7 | GO:1900620 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.1 | 0.4 | GO:0019062 | virion attachment to host cell(GO:0019062) adhesion of symbiont to host(GO:0044406) adhesion of symbiont to host cell(GO:0044650) |
| 0.1 | 0.9 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 0.4 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.1 | 0.4 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 0.4 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.1 | 0.2 | GO:0035971 | peptidyl-histidine dephosphorylation(GO:0035971) |
| 0.1 | 0.2 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.1 | 0.4 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.4 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.1 | 0.2 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) L-kynurenine catabolic process(GO:0097053) |
| 0.1 | 0.2 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.1 | 0.2 | GO:0099548 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.1 | 0.2 | GO:1903385 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 0.2 | GO:0090076 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 0.1 | 0.2 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 0.2 | GO:0060547 | negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.0 | 0.5 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.1 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.5 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.0 | 0.8 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.2 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.2 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.4 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.2 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.1 | GO:1902514 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.0 | 0.2 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 0.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.0 | 0.2 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.2 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.2 | GO:0019430 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 3.0 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.1 | GO:0043584 | nose development(GO:0043584) |
| 0.0 | 0.3 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.2 | GO:0060005 | reflex(GO:0060004) vestibular reflex(GO:0060005) |
| 0.0 | 0.2 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.9 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.1 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.0 | 0.5 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.3 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.1 | GO:0048313 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.0 | 0.7 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.3 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.1 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.0 | 0.1 | GO:0070285 | pigment cell development(GO:0070285) |
| 0.0 | 0.0 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.1 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.0 | 0.6 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.2 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 | 0.1 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.3 | GO:0006415 | translational termination(GO:0006415) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.2 | GO:0021654 | rhombomere boundary formation(GO:0021654) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.1 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.2 | GO:0042119 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 0.0 | 0.1 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.1 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.0 | 0.0 | GO:0071706 | tumor necrosis factor production(GO:0032640) tumor necrosis factor superfamily cytokine production(GO:0071706) |
| 0.0 | 0.0 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.0 | 0.1 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.5 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 0.1 | GO:0060386 | synapse assembly involved in innervation(GO:0060386) |
| 0.0 | 0.1 | GO:0050909 | sensory perception of taste(GO:0050909) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 0.2 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.4 | GO:0071914 | prominosome(GO:0071914) |
| 0.0 | 0.1 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.0 | 0.1 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.0 | 0.6 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.9 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.2 | GO:1990246 | uniplex complex(GO:1990246) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.1 | 0.4 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.1 | 0.4 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.1 | 0.4 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.1 | 0.4 | GO:0034618 | arginine binding(GO:0034618) |
| 0.1 | 0.3 | GO:0015462 | protein-transmembrane transporting ATPase activity(GO:0015462) efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 0.3 | GO:0001635 | adrenomedullin receptor activity(GO:0001605) calcitonin gene-related peptide receptor activity(GO:0001635) |
| 0.1 | 0.5 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.1 | 0.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 0.4 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.2 | GO:0101006 | protein histidine phosphatase activity(GO:0101006) |
| 0.1 | 0.3 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.1 | 0.7 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.2 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.0 | 0.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.2 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.2 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.4 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.2 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.4 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 2.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.6 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.3 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0004134 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.0 | 0.1 | GO:0102344 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.0 | 0.4 | GO:0008308 | voltage-gated chloride channel activity(GO:0005247) voltage-gated anion channel activity(GO:0008308) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.2 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.1 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.0 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.1 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.0 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.0 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.3 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.7 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.2 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.1 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.1 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |