Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
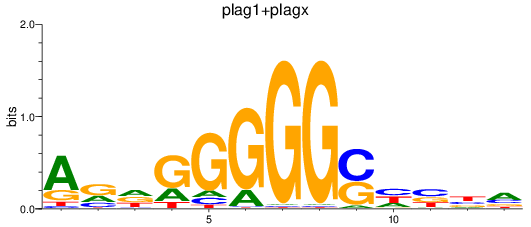
Results for plag1+plagx
Z-value: 0.94
Transcription factors associated with plag1+plagx
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
plagx
|
ENSDARG00000036855 | pleiomorphic adenoma gene X |
|
plag1
|
ENSDARG00000051926 | pleiomorphic adenoma gene 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| plagx | dr11_v1_chr23_+_32011768_32011768 | -0.72 | 4.8e-04 | Click! |
| plag1 | dr11_v1_chr7_+_58736889_58736889 | 0.47 | 4.1e-02 | Click! |
Activity profile of plag1+plagx motif
Sorted Z-values of plag1+plagx motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_22310919 | 8.71 |
ENSDART00000108719
|
crygm2d10
|
crystallin, gamma M2d10 |
| chr1_+_49568335 | 4.78 |
ENSDART00000142957
|
col17a1a
|
collagen, type XVII, alpha 1a |
| chr8_-_25247284 | 3.74 |
ENSDART00000132697
|
gnat2
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2 |
| chr7_+_568819 | 3.65 |
ENSDART00000173716
|
nrxn2b
|
neurexin 2b |
| chr19_-_3193912 | 3.24 |
ENSDART00000133159
|
si:ch211-133n4.6
|
si:ch211-133n4.6 |
| chr12_+_16440708 | 3.16 |
ENSDART00000113810
|
ankrd1b
|
ankyrin repeat domain 1b (cardiac muscle) |
| chr12_-_25916530 | 2.74 |
ENSDART00000186386
|
sncgb
|
synuclein, gamma b (breast cancer-specific protein 1) |
| chr16_-_36798783 | 2.45 |
ENSDART00000145697
|
calb1
|
calbindin 1 |
| chr24_-_26383978 | 2.04 |
ENSDART00000031426
|
skilb
|
SKI-like proto-oncogene b |
| chr7_-_38477235 | 1.79 |
ENSDART00000084355
|
zgc:165481
|
zgc:165481 |
| chr4_+_19534833 | 1.73 |
ENSDART00000140028
|
lrrc4.1
|
leucine rich repeat containing 4.1 |
| chr2_+_51783120 | 1.48 |
ENSDART00000177559
|
crygn1
|
crystallin, gamma N1 |
| chr20_-_14665002 | 1.41 |
ENSDART00000152816
|
scrn2
|
secernin 2 |
| chr4_+_9400012 | 1.26 |
ENSDART00000191960
|
tmtc1
|
transmembrane and tetratricopeptide repeat containing 1 |
| chr4_+_6572364 | 1.25 |
ENSDART00000122574
|
ppp1r3aa
|
protein phosphatase 1, regulatory subunit 3Aa |
| chr24_+_35564668 | 1.24 |
ENSDART00000122734
|
cebpd
|
CCAAT/enhancer binding protein (C/EBP), delta |
| chr14_-_2209742 | 1.20 |
ENSDART00000054889
|
pcdh2ab5
|
protocadherin 2 alpha b 5 |
| chr1_+_47446032 | 1.19 |
ENSDART00000126904
ENSDART00000007262 |
gja8b
|
gap junction protein alpha 8 paralog b |
| chr19_-_8940068 | 1.17 |
ENSDART00000043507
|
ciarta
|
circadian associated repressor of transcription a |
| chr23_+_44732863 | 1.13 |
ENSDART00000160044
ENSDART00000172268 |
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr22_-_4798738 | 0.92 |
ENSDART00000114465
|
zgc:195170
|
zgc:195170 |
| chr8_+_25247245 | 0.91 |
ENSDART00000045798
|
ampd2b
|
adenosine monophosphate deaminase 2b |
| chr6_-_35401282 | 0.86 |
ENSDART00000127612
|
rgs5a
|
regulator of G protein signaling 5a |
| chr21_+_7823146 | 0.81 |
ENSDART00000030579
|
crhbp
|
corticotropin releasing hormone binding protein |
| chr12_+_22657925 | 0.80 |
ENSDART00000153048
|
si:dkey-219e21.4
|
si:dkey-219e21.4 |
| chr2_+_13069168 | 0.79 |
ENSDART00000192832
|
prkag2b
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit b |
| chr19_+_43684376 | 0.79 |
ENSDART00000051723
|
si:ch211-193k19.1
|
si:ch211-193k19.1 |
| chr7_-_41403022 | 0.79 |
ENSDART00000174285
|
BX322787.1
|
|
| chr24_-_21490628 | 0.68 |
ENSDART00000181546
|
atp8a2
|
ATPase phospholipid transporting 8A2 |
| chr22_+_8551813 | 0.67 |
ENSDART00000129690
|
zmp:0000000984
|
zmp:0000000984 |
| chr12_-_46112892 | 0.66 |
ENSDART00000187128
ENSDART00000114268 |
zgc:153932
|
zgc:153932 |
| chr19_-_32944050 | 0.63 |
ENSDART00000137611
|
azin1b
|
antizyme inhibitor 1b |
| chr5_-_23878809 | 0.62 |
ENSDART00000160603
|
si:ch211-135f11.4
|
si:ch211-135f11.4 |
| chr16_+_11834516 | 0.49 |
ENSDART00000146611
|
cxcr3.3
|
chemokine (C-X-C motif) receptor 3, tandem duplicate 3 |
| chr12_-_30345444 | 0.40 |
ENSDART00000152985
|
vwa2
|
von Willebrand factor A domain containing 2 |
| chr10_-_36240806 | 0.37 |
ENSDART00000171468
|
or109-2
|
odorant receptor, family D, subfamily 109, member 2 |
| chr23_+_30859086 | 0.37 |
ENSDART00000053661
|
zgc:198371
|
zgc:198371 |
| chr3_+_18795570 | 0.34 |
ENSDART00000042368
|
fahd1
|
fumarylacetoacetate hydrolase domain containing 1 |
| chr14_-_1958994 | 0.26 |
ENSDART00000161783
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr14_-_33177935 | 0.19 |
ENSDART00000180583
ENSDART00000078856 |
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr6_+_52911338 | 0.17 |
ENSDART00000065710
|
si:dkeyp-3f10.14
|
si:dkeyp-3f10.14 |
| chr19_+_37620342 | 0.14 |
ENSDART00000158960
|
thsd7aa
|
thrombospondin, type I, domain containing 7Aa |
| chr9_+_22466218 | 0.08 |
ENSDART00000131214
|
dgkg
|
diacylglycerol kinase, gamma |
| chr19_+_11260143 | 0.03 |
ENSDART00000187771
|
si:ch73-171o20.1
|
si:ch73-171o20.1 |
| chr21_+_5801105 | 0.01 |
ENSDART00000151225
ENSDART00000184487 |
ccng2
|
cyclin G2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of plag1+plagx
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.4 | GO:0099509 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.4 | 3.7 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.3 | 0.8 | GO:0002792 | negative regulation of peptide secretion(GO:0002792) negative regulation of peptide hormone secretion(GO:0090278) |
| 0.1 | 2.7 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.1 | 0.6 | GO:1902269 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.1 | 2.0 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.1 | 0.9 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 8.6 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.1 | 1.3 | GO:0010962 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.1 | 1.1 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 4.8 | GO:0008544 | epidermis development(GO:0008544) |
| 0.0 | 1.2 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.8 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.7 | GO:0045332 | phospholipid translocation(GO:0045332) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.7 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 5.2 | GO:0043679 | axon terminus(GO:0043679) |
| 0.1 | 1.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.1 | 1.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 1.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.8 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 5.2 | GO:0031012 | extracellular matrix(GO:0031012) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.8 | 2.4 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.8 | 3.2 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.3 | 1.4 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.2 | 1.2 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.1 | 10.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 3.7 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 0.6 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.1 | 1.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 1.3 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 0.9 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.3 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.1 | 1.1 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 5.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 2.0 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.8 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.8 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 0.5 | GO:0019957 | C-C chemokine binding(GO:0019957) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.7 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.2 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.7 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 1.2 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |