Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
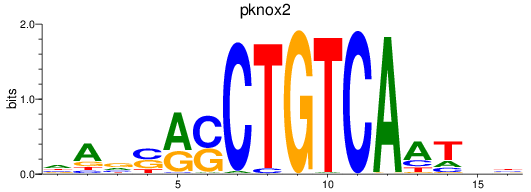
Results for pknox2
Z-value: 0.91
Transcription factors associated with pknox2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pknox2
|
ENSDARG00000055349 | pbx/knotted 1 homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pknox2 | dr11_v1_chr10_-_31220558_31220558 | -0.25 | 2.9e-01 | Click! |
Activity profile of pknox2 motif
Sorted Z-values of pknox2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_26288301 | 3.15 |
ENSDART00000017668
|
ptbp1a
|
polypyrimidine tract binding protein 1a |
| chr15_-_20528494 | 2.85 |
ENSDART00000048423
|
timm50
|
translocase of inner mitochondrial membrane 50 homolog (S. cerevisiae) |
| chr3_-_10312232 | 2.75 |
ENSDART00000081766
|
nog1
|
noggin 1 |
| chr8_-_18613948 | 2.49 |
ENSDART00000089172
|
cpox
|
coproporphyrinogen oxidase |
| chr14_-_41468892 | 2.25 |
ENSDART00000173099
ENSDART00000003170 |
mid1ip1l
|
MID1 interacting protein 1, like |
| chr23_-_31060350 | 2.18 |
ENSDART00000145598
ENSDART00000191491 |
si:ch211-197l9.5
|
si:ch211-197l9.5 |
| chr20_-_28433990 | 2.17 |
ENSDART00000182824
ENSDART00000193381 |
wdr21
|
WD repeat domain 21 |
| chr3_+_28576173 | 2.07 |
ENSDART00000151189
|
sept12
|
septin 12 |
| chr12_-_48168135 | 2.06 |
ENSDART00000186624
|
pald1a
|
phosphatase domain containing, paladin 1a |
| chr6_+_14949950 | 2.02 |
ENSDART00000149202
ENSDART00000149949 |
pou3f3b
|
POU class 3 homeobox 3b |
| chr16_+_40575742 | 1.93 |
ENSDART00000161503
|
ccne2
|
cyclin E2 |
| chr22_+_25086942 | 1.92 |
ENSDART00000061117
|
rrbp1b
|
ribosome binding protein 1b |
| chr2_+_45081489 | 1.92 |
ENSDART00000123966
|
chrng
|
cholinergic receptor, nicotinic, gamma |
| chr18_+_24562188 | 1.89 |
ENSDART00000099463
|
lysmd4
|
LysM, putative peptidoglycan-binding, domain containing 4 |
| chr17_-_7792376 | 1.87 |
ENSDART00000064655
|
zbtb2a
|
zinc finger and BTB domain containing 2a |
| chr22_+_25086567 | 1.83 |
ENSDART00000192114
ENSDART00000177284 ENSDART00000180296 ENSDART00000190384 |
rrbp1b
|
ribosome binding protein 1b |
| chr4_+_12931763 | 1.76 |
ENSDART00000016382
|
wif1
|
wnt inhibitory factor 1 |
| chr23_+_37815645 | 1.75 |
ENSDART00000011627
|
irx7
|
iroquois homeobox 7 |
| chr16_-_13595027 | 1.74 |
ENSDART00000060004
|
ntd5
|
ntl-dependent gene 5 |
| chr4_-_20081621 | 1.65 |
ENSDART00000024647
|
dennd6b
|
DENN/MADD domain containing 6B |
| chr5_-_12219572 | 1.64 |
ENSDART00000167834
|
nos1
|
nitric oxide synthase 1 (neuronal) |
| chr10_+_42521943 | 1.59 |
ENSDART00000010420
ENSDART00000075303 |
actr1
|
ARP1 actin related protein 1, centractin |
| chr15_+_36309070 | 1.58 |
ENSDART00000157034
|
gmnc
|
geminin coiled-coil domain containing |
| chr8_-_4760723 | 1.57 |
ENSDART00000064201
|
cdc45
|
CDC45 cell division cycle 45 homolog (S. cerevisiae) |
| chr2_+_19195841 | 1.55 |
ENSDART00000163137
ENSDART00000161095 |
elovl1a
|
ELOVL fatty acid elongase 1a |
| chr25_+_3549401 | 1.54 |
ENSDART00000166312
|
ccdc77
|
coiled-coil domain containing 77 |
| chr12_+_36971952 | 1.51 |
ENSDART00000125900
|
hs3st3b1b
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1b |
| chr17_+_23964132 | 1.50 |
ENSDART00000154823
|
xpo1b
|
exportin 1 (CRM1 homolog, yeast) b |
| chr20_+_32478151 | 1.43 |
ENSDART00000145269
|
ostm1
|
osteopetrosis associated transmembrane protein 1 |
| chr19_+_25465025 | 1.39 |
ENSDART00000018553
|
rpa3
|
replication protein A3 |
| chr19_+_18799319 | 1.36 |
ENSDART00000171843
|
ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr8_-_17184482 | 1.36 |
ENSDART00000025803
|
pola2
|
polymerase (DNA directed), alpha 2 |
| chr20_-_20270191 | 1.35 |
ENSDART00000009356
|
ppp2r5ea
|
protein phosphatase 2, regulatory subunit B', epsilon isoform a |
| chr22_-_37834312 | 1.34 |
ENSDART00000076128
|
ppp1r2
|
protein phosphatase 1, regulatory (inhibitor) subunit 2 |
| chr19_-_15281996 | 1.34 |
ENSDART00000103784
|
edn2
|
endothelin 2 |
| chr7_-_28565230 | 1.32 |
ENSDART00000028887
|
tmem9b
|
TMEM9 domain family, member B |
| chr7_-_26262978 | 1.30 |
ENSDART00000137769
|
ap1s1
|
adaptor-related protein complex 1, sigma 1 subunit |
| chr16_-_7239457 | 1.25 |
ENSDART00000148992
ENSDART00000149260 |
nt5c3a
|
5'-nucleotidase, cytosolic IIIA |
| chr11_-_15874974 | 1.24 |
ENSDART00000166551
ENSDART00000129526 ENSDART00000165836 |
rap1ab
|
RAP1A, member of RAS oncogene family b |
| chr7_+_57089354 | 1.24 |
ENSDART00000140702
|
scamp2l
|
secretory carrier membrane protein 2, like |
| chr10_+_25982212 | 1.22 |
ENSDART00000128292
ENSDART00000108808 |
frem2a
|
Fras1 related extracellular matrix protein 2a |
| chr2_-_11027258 | 1.20 |
ENSDART00000081072
ENSDART00000193824 ENSDART00000187036 ENSDART00000097741 |
ssbp3a
|
single stranded DNA binding protein 3a |
| chr20_-_28433616 | 1.19 |
ENSDART00000169289
|
wdr21
|
WD repeat domain 21 |
| chr21_+_26539157 | 1.17 |
ENSDART00000021121
|
stx5al
|
syntaxin 5A, like |
| chr2_-_45510699 | 1.17 |
ENSDART00000024034
ENSDART00000145634 |
gpsm2
|
G protein signaling modulator 2 |
| chr19_-_30800004 | 1.16 |
ENSDART00000128560
ENSDART00000045504 ENSDART00000125893 |
trit1
|
tRNA isopentenyltransferase 1 |
| chr16_-_52646789 | 1.14 |
ENSDART00000035761
|
ubr5
|
ubiquitin protein ligase E3 component n-recognin 5 |
| chr12_+_18899396 | 1.12 |
ENSDART00000105858
|
xrcc6
|
X-ray repair complementing defective repair in Chinese hamster cells 6 |
| chr21_-_34658266 | 1.12 |
ENSDART00000023038
|
dacha
|
dachshund a |
| chr11_-_10456387 | 1.10 |
ENSDART00000011087
ENSDART00000081827 |
ect2
|
epithelial cell transforming 2 |
| chr10_+_20590190 | 1.07 |
ENSDART00000131819
|
letm2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr14_+_22022441 | 1.07 |
ENSDART00000149121
|
clcf1
|
cardiotrophin-like cytokine factor 1 |
| chr2_-_24402341 | 1.05 |
ENSDART00000155442
ENSDART00000088572 |
zgc:154006
|
zgc:154006 |
| chr8_+_30425700 | 1.00 |
ENSDART00000012132
|
cbwd
|
COBW domain containing |
| chr18_+_17786548 | 0.98 |
ENSDART00000189493
ENSDART00000146133 |
ZNF423
|
si:ch211-216l23.1 |
| chr24_+_29352039 | 0.97 |
ENSDART00000101641
|
prmt6
|
protein arginine methyltransferase 6 |
| chr21_-_38153824 | 0.97 |
ENSDART00000151226
|
klf5l
|
Kruppel-like factor 5 like |
| chr5_-_17601759 | 0.96 |
ENSDART00000138387
|
si:ch211-130h14.6
|
si:ch211-130h14.6 |
| chr8_+_25959940 | 0.96 |
ENSDART00000143011
ENSDART00000140626 |
si:dkey-72l14.4
|
si:dkey-72l14.4 |
| chr24_+_33392698 | 0.95 |
ENSDART00000122579
|
si:ch73-173p19.1
|
si:ch73-173p19.1 |
| chr18_+_17786710 | 0.95 |
ENSDART00000190203
ENSDART00000187095 ENSDART00000083296 |
ZNF423
|
si:ch211-216l23.1 |
| chr17_-_14780578 | 0.94 |
ENSDART00000154690
|
si:ch211-266o15.1
|
si:ch211-266o15.1 |
| chr7_-_26263183 | 0.93 |
ENSDART00000079357
ENSDART00000190369 ENSDART00000193154 ENSDART00000101109 |
ap1s1
|
adaptor-related protein complex 1, sigma 1 subunit |
| chr23_-_9768700 | 0.92 |
ENSDART00000045126
|
lama5
|
laminin, alpha 5 |
| chr21_+_43882274 | 0.92 |
ENSDART00000075672
|
sra1
|
steroid receptor RNA activator 1 |
| chr7_-_56766100 | 0.90 |
ENSDART00000189934
|
csnk2a2a
|
casein kinase 2, alpha prime polypeptide a |
| chr6_+_41452979 | 0.90 |
ENSDART00000007353
|
wdr82
|
WD repeat domain 82 |
| chr10_+_20589969 | 0.88 |
ENSDART00000183042
|
letm2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr9_+_34151367 | 0.87 |
ENSDART00000143991
|
gpr161
|
G protein-coupled receptor 161 |
| chr4_+_25693463 | 0.87 |
ENSDART00000132864
|
acot18
|
acyl-CoA thioesterase 18 |
| chr12_-_17201028 | 0.86 |
ENSDART00000020541
|
lipf
|
lipase, gastric |
| chr13_-_42749916 | 0.84 |
ENSDART00000140019
|
capn2a
|
calpain 2, (m/II) large subunit a |
| chr17_-_36860988 | 0.83 |
ENSDART00000154981
|
senp6b
|
SUMO1/sentrin specific peptidase 6b |
| chr16_+_13855039 | 0.82 |
ENSDART00000113764
ENSDART00000143983 |
zgc:174888
|
zgc:174888 |
| chr20_+_47434709 | 0.81 |
ENSDART00000067776
|
rab10
|
RAB10, member RAS oncogene family |
| chr23_+_1661743 | 0.81 |
ENSDART00000044776
|
stxbp3
|
syntaxin binding protein 3 |
| chr11_+_44503774 | 0.80 |
ENSDART00000169295
|
ero1b
|
endoplasmic reticulum oxidoreductase beta |
| chr25_+_14870043 | 0.80 |
ENSDART00000035714
ENSDART00000171835 |
dnajc24
|
DnaJ (Hsp40) homolog, subfamily C, member 24 |
| chr22_-_21046654 | 0.80 |
ENSDART00000064902
|
ssbp4
|
single stranded DNA binding protein 4 |
| chr22_+_2417105 | 0.80 |
ENSDART00000106415
|
zgc:113220
|
zgc:113220 |
| chr4_-_16345227 | 0.79 |
ENSDART00000079521
|
kera
|
keratocan |
| chr11_-_843811 | 0.78 |
ENSDART00000173331
|
atg7
|
ATG7 autophagy related 7 homolog (S. cerevisiae) |
| chr23_+_18103080 | 0.76 |
ENSDART00000010270
|
mfsd4ab
|
major facilitator superfamily domain containing 4Ab |
| chr20_+_21391181 | 0.74 |
ENSDART00000185158
ENSDART00000049586 ENSDART00000024922 |
jag2b
|
jagged 2b |
| chr22_-_21046843 | 0.74 |
ENSDART00000133982
|
ssbp4
|
single stranded DNA binding protein 4 |
| chr2_+_15128418 | 0.73 |
ENSDART00000141921
|
arhgap29b
|
Rho GTPase activating protein 29b |
| chr5_-_69437422 | 0.70 |
ENSDART00000073676
|
isca1
|
iron-sulfur cluster assembly 1 |
| chr23_-_24263474 | 0.70 |
ENSDART00000160312
|
hspb7
|
heat shock protein family, member 7 (cardiovascular) |
| chr12_+_3871452 | 0.69 |
ENSDART00000066546
|
nif3l1
|
NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae) |
| chr1_-_19431510 | 0.68 |
ENSDART00000089025
|
lap3
|
leucine aminopeptidase 3 |
| chr22_-_17677947 | 0.67 |
ENSDART00000139911
|
tjp3
|
tight junction protein 3 |
| chr5_+_22307605 | 0.67 |
ENSDART00000138154
|
arhgap20b
|
Rho GTPase activating protein 20b |
| chr19_+_42693855 | 0.67 |
ENSDART00000136873
|
clasp2
|
cytoplasmic linker associated protein 2 |
| chr19_-_9712530 | 0.67 |
ENSDART00000134816
|
slc2a3a
|
solute carrier family 2 (facilitated glucose transporter), member 3a |
| chr14_+_29941266 | 0.65 |
ENSDART00000112757
|
fam149a
|
family with sequence similarity 149 member A |
| chr22_+_18316144 | 0.65 |
ENSDART00000137985
|
gatad2ab
|
GATA zinc finger domain containing 2Ab |
| chr13_+_33655404 | 0.64 |
ENSDART00000023379
|
mgme1
|
mitochondrial genome maintenance exonuclease 1 |
| chr8_+_21437908 | 0.64 |
ENSDART00000142758
|
si:dkey-163f12.10
|
si:dkey-163f12.10 |
| chr7_+_20467549 | 0.64 |
ENSDART00000173724
|
si:dkey-33c9.8
|
si:dkey-33c9.8 |
| chr8_-_32385989 | 0.61 |
ENSDART00000143716
ENSDART00000098850 |
lipg
|
lipase, endothelial |
| chr15_+_23528010 | 0.61 |
ENSDART00000152786
|
si:dkey-182i3.8
|
si:dkey-182i3.8 |
| chr22_-_20720427 | 0.61 |
ENSDART00000105532
|
oaz1a
|
ornithine decarboxylase antizyme 1a |
| chr25_+_19008497 | 0.60 |
ENSDART00000104420
|
samm50
|
SAMM50 sorting and assembly machinery component |
| chr17_-_23673864 | 0.59 |
ENSDART00000104738
ENSDART00000128958 |
ptena
|
phosphatase and tensin homolog A |
| chr19_+_43905671 | 0.59 |
ENSDART00000168725
ENSDART00000133628 |
ankib1a
|
ankyrin repeat and IBR domain containing 1a |
| chr9_-_12885201 | 0.58 |
ENSDART00000124957
|
ankzf1
|
ankyrin repeat and zinc finger domain containing 1 |
| chr2_+_30489846 | 0.58 |
ENSDART00000145732
|
march6
|
membrane-associated ring finger (C3HC4) 6 |
| chr11_-_19598809 | 0.57 |
ENSDART00000110499
|
atxn7
|
ataxin 7 |
| chr15_+_23528310 | 0.57 |
ENSDART00000152523
|
si:dkey-182i3.8
|
si:dkey-182i3.8 |
| chr5_-_60159116 | 0.57 |
ENSDART00000147675
|
si:dkey-280e8.1
|
si:dkey-280e8.1 |
| chr10_+_28160265 | 0.55 |
ENSDART00000022484
|
rnft1
|
ring finger protein, transmembrane 1 |
| chr3_-_20793655 | 0.53 |
ENSDART00000163473
ENSDART00000159457 |
spop
|
speckle-type POZ protein |
| chr19_+_32553874 | 0.53 |
ENSDART00000078197
|
heyl
|
hes-related family bHLH transcription factor with YRPW motif-like |
| chr5_-_32489796 | 0.52 |
ENSDART00000168870
|
gpr107
|
G protein-coupled receptor 107 |
| chr20_-_46114467 | 0.50 |
ENSDART00000126495
|
taar12i
|
trace amine associated receptor 12i |
| chr20_+_27712714 | 0.50 |
ENSDART00000008306
|
zbtb1
|
zinc finger and BTB domain containing 1 |
| chr23_+_25893020 | 0.50 |
ENSDART00000144769
|
pkig
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr11_+_5880562 | 0.49 |
ENSDART00000129663
ENSDART00000130768 ENSDART00000160909 |
dazap1
|
DAZ associated protein 1 |
| chr15_-_28587147 | 0.48 |
ENSDART00000156049
|
ssh2a
|
slingshot protein phosphatase 2a |
| chr10_+_11355841 | 0.47 |
ENSDART00000193067
ENSDART00000064215 |
cops4
|
COP9 constitutive photomorphogenic homolog subunit 4 (Arabidopsis) |
| chr3_-_40836081 | 0.47 |
ENSDART00000143135
|
wipi2
|
WD repeat domain, phosphoinositide interacting 2 |
| chr7_-_19369002 | 0.46 |
ENSDART00000165680
|
ntn4
|
netrin 4 |
| chr12_+_28854963 | 0.46 |
ENSDART00000153227
|
nfe2l1b
|
nuclear factor, erythroid 2-like 1b |
| chr7_-_37555208 | 0.45 |
ENSDART00000148905
ENSDART00000150229 |
cylda
|
cylindromatosis (turban tumor syndrome), a |
| chr9_+_27876146 | 0.45 |
ENSDART00000133997
|
armc8
|
armadillo repeat containing 8 |
| chr7_+_52761841 | 0.45 |
ENSDART00000111444
|
ppip5k1a
|
diphosphoinositol pentakisphosphate kinase 1a |
| chr9_-_33328948 | 0.44 |
ENSDART00000006948
|
rpl8
|
ribosomal protein L8 |
| chr20_+_329032 | 0.44 |
ENSDART00000036635
|
fynb
|
FYN proto-oncogene, Src family tyrosine kinase b |
| chr9_-_48281941 | 0.44 |
ENSDART00000099787
|
klhl41a
|
kelch-like family member 41a |
| chr10_+_36695597 | 0.42 |
ENSDART00000169015
ENSDART00000171392 |
rab6a
|
RAB6A, member RAS oncogene family |
| chr9_-_24970018 | 0.42 |
ENSDART00000026924
|
dnah7
|
dynein, axonemal, heavy chain 7 |
| chr13_-_7233811 | 0.42 |
ENSDART00000162026
|
ninl
|
ninein-like |
| chr3_-_36260102 | 0.41 |
ENSDART00000126588
|
rac3a
|
Rac family small GTPase 3a |
| chr19_-_81477 | 0.40 |
ENSDART00000159815
|
hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr5_-_41124241 | 0.39 |
ENSDART00000083561
|
mtmr12
|
myotubularin related protein 12 |
| chr17_-_51679222 | 0.39 |
ENSDART00000155062
|
atp6v1c2
|
ATPase H+ transporting V1 subunit C2 |
| chr2_-_55861351 | 0.38 |
ENSDART00000059003
|
rx2
|
retinal homeobox gene 2 |
| chr21_+_38312549 | 0.35 |
ENSDART00000065159
|
zgc:158291
|
zgc:158291 |
| chr13_+_35765317 | 0.35 |
ENSDART00000100156
ENSDART00000167650 |
agpat4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta) |
| chr24_+_10413484 | 0.34 |
ENSDART00000111014
|
myca
|
MYC proto-oncogene, bHLH transcription factor a |
| chr23_+_3734491 | 0.34 |
ENSDART00000185982
|
smim29
|
small integral membrane protein 29 |
| chr9_-_30274412 | 0.33 |
ENSDART00000089526
|
otc
|
ornithine carbamoyltransferase |
| chr17_-_43666166 | 0.33 |
ENSDART00000077990
|
egr2a
|
early growth response 2a |
| chr7_+_30725473 | 0.31 |
ENSDART00000085716
|
mtmr10
|
myotubularin related protein 10 |
| chr8_+_45294767 | 0.30 |
ENSDART00000191527
|
ubap2b
|
ubiquitin associated protein 2b |
| chr1_-_19402802 | 0.30 |
ENSDART00000135552
|
rbm47
|
RNA binding motif protein 47 |
| chr7_-_56766973 | 0.30 |
ENSDART00000020967
|
csnk2a2a
|
casein kinase 2, alpha prime polypeptide a |
| chr22_-_22231720 | 0.30 |
ENSDART00000160165
|
ap3d1
|
adaptor-related protein complex 3, delta 1 subunit |
| chr18_+_29402623 | 0.30 |
ENSDART00000014703
|
mafa
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog a (paralog a) |
| chr5_+_72145468 | 0.29 |
ENSDART00000148626
|
abl1
|
c-abl oncogene 1, non-receptor tyrosine kinase |
| chr9_+_33267211 | 0.29 |
ENSDART00000025635
|
usp9
|
ubiquitin specific peptidase 9 |
| chr11_+_28218141 | 0.28 |
ENSDART00000043756
|
ephb2b
|
eph receptor B2b |
| chr24_-_21511737 | 0.28 |
ENSDART00000109587
|
rnf6
|
ring finger protein (C3H2C3 type) 6 |
| chr5_+_57743815 | 0.27 |
ENSDART00000005090
|
alg9
|
ALG9, alpha-1,2-mannosyltransferase |
| chr4_+_16787488 | 0.27 |
ENSDART00000143006
|
golt1ba
|
golgi transport 1Ba |
| chr21_-_588858 | 0.26 |
ENSDART00000168983
|
TMEM38B
|
transmembrane protein 38B |
| chr6_+_53288018 | 0.25 |
ENSDART00000170319
|
cysltr3
|
cysteinyl leukotriene receptor 3 |
| chr1_+_57145072 | 0.19 |
ENSDART00000152776
|
si:ch73-94k4.4
|
si:ch73-94k4.4 |
| chr20_-_7547080 | 0.18 |
ENSDART00000146135
|
usp24
|
ubiquitin specific peptidase 24 |
| chr3_-_23596532 | 0.18 |
ENSDART00000124921
|
ube2z
|
ubiquitin-conjugating enzyme E2Z |
| chr23_+_24705424 | 0.14 |
ENSDART00000104029
|
c1qtnf12
|
C1q and TNF related 12 |
| chr13_+_28417297 | 0.14 |
ENSDART00000043658
|
slc2a15a
|
solute carrier family 2 (facilitated glucose transporter), member 15a |
| chr3_-_25646149 | 0.14 |
ENSDART00000122735
|
usp43b
|
ubiquitin specific peptidase 43b |
| chr6_-_23931442 | 0.13 |
ENSDART00000160547
|
sec16b
|
SEC16 homolog B, endoplasmic reticulum export factor |
| chr3_-_15889508 | 0.07 |
ENSDART00000148363
|
cramp1
|
cramped chromatin regulator homolog 1 |
| chr8_+_24745041 | 0.06 |
ENSDART00000148872
|
slc16a4
|
solute carrier family 16, member 4 |
| chr12_-_44307963 | 0.06 |
ENSDART00000161009
|
si:ch73-329n5.1
|
si:ch73-329n5.1 |
| chr10_+_37500234 | 0.06 |
ENSDART00000132096
ENSDART00000099473 |
msi2a
|
musashi RNA-binding protein 2a |
| chr25_-_8138122 | 0.05 |
ENSDART00000104659
|
sergef
|
secretion regulating guanine nucleotide exchange factor |
| chr12_-_36740781 | 0.05 |
ENSDART00000105484
|
si:ch211-216b21.2
|
si:ch211-216b21.2 |
| chr3_-_16055432 | 0.05 |
ENSDART00000123621
ENSDART00000023859 |
atp6v0ca
|
ATPase H+ transporting V0 subunit ca |
| chr16_-_563732 | 0.05 |
ENSDART00000183394
|
irx2a
|
iroquois homeobox 2a |
| chr25_+_3549584 | 0.04 |
ENSDART00000165913
|
ccdc77
|
coiled-coil domain containing 77 |
| chr8_+_25616946 | 0.02 |
ENSDART00000133983
|
slc38a5a
|
solute carrier family 38, member 5a |
| chr20_+_26690036 | 0.02 |
ENSDART00000103232
|
foxf2b
|
forkhead box F2b |
| chr14_+_30340251 | 0.01 |
ENSDART00000148448
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr12_-_36740306 | 0.00 |
ENSDART00000153259
|
si:ch211-216b21.2
|
si:ch211-216b21.2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of pknox2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.1 | GO:0060467 | negative regulation of fertilization(GO:0060467) prevention of polyspermy(GO:0060468) cortical granule exocytosis(GO:0060471) |
| 0.5 | 1.6 | GO:0098924 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) retrograde trans-synaptic signaling by nitric oxide(GO:0098924) |
| 0.3 | 1.4 | GO:0045428 | regulation of nitric oxide biosynthetic process(GO:0045428) positive regulation of nitric oxide biosynthetic process(GO:0045429) positive regulation of reactive oxygen species biosynthetic process(GO:1903428) positive regulation of nitric oxide metabolic process(GO:1904407) |
| 0.3 | 1.6 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.3 | 0.9 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.3 | 1.3 | GO:0060585 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.3 | 2.8 | GO:0042664 | negative regulation of endodermal cell fate specification(GO:0042664) |
| 0.2 | 1.2 | GO:0051591 | negative regulation of neurotransmitter secretion(GO:0046929) response to cAMP(GO:0051591) cellular response to cAMP(GO:0071320) negative regulation of synaptic vesicle transport(GO:1902804) negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.2 | 1.0 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.2 | 1.1 | GO:0071480 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.2 | 0.6 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.2 | 0.8 | GO:0090234 | regulation of centromere complex assembly(GO:0090230) regulation of kinetochore assembly(GO:0090234) |
| 0.2 | 0.6 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.2 | 1.1 | GO:1902914 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.2 | 1.8 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.2 | 1.6 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.2 | 1.5 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.2 | 2.8 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.2 | 2.5 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.2 | 1.1 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.1 | 0.8 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.6 | GO:0031174 | lifelong otolith mineralization(GO:0031174) |
| 0.1 | 0.8 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 0.6 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
| 0.1 | 0.7 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 0.7 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 0.1 | 0.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 1.9 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.1 | 1.1 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.1 | 0.4 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.1 | 0.5 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.3 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 3.3 | GO:0048794 | swim bladder development(GO:0048794) |
| 0.1 | 0.5 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.1 | 0.8 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.1 | 0.7 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.1 | 1.3 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.1 | 1.0 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.1 | 1.4 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.1 | 1.2 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.1 | 1.3 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 | 0.6 | GO:0021684 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.3 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.9 | GO:0035141 | medial fin morphogenesis(GO:0035141) |
| 0.0 | 1.4 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 2.2 | GO:0071166 | ribonucleoprotein complex localization(GO:0071166) |
| 0.0 | 0.7 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.6 | GO:0006596 | polyamine biosynthetic process(GO:0006596) |
| 0.0 | 0.5 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 1.9 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.4 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 1.6 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.6 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.1 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.0 | 1.2 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.0 | 0.7 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.5 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.1 | GO:0070863 | regulation of protein exit from endoplasmic reticulum(GO:0070861) positive regulation of protein exit from endoplasmic reticulum(GO:0070863) protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 1.1 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.4 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 2.1 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 1.2 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 1.2 | GO:0033334 | fin morphogenesis(GO:0033334) |
| 0.0 | 0.4 | GO:0071910 | determination of liver left/right asymmetry(GO:0071910) |
| 0.0 | 0.5 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 1.2 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 1.2 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.6 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.6 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.3 | 1.4 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.3 | 2.8 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.2 | 1.6 | GO:0031261 | DNA replication preinitiation complex(GO:0031261) |
| 0.2 | 1.4 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.2 | 1.9 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 1.2 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.1 | 0.4 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 0.8 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 0.6 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 1.4 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.1 | 1.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 2.5 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 0.5 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.4 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.1 | 2.1 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 0.9 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.5 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 5.9 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.8 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.6 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.7 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 2.5 | GO:0048475 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.0 | 1.6 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.6 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.6 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.1 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.4 | GO:0016529 | sarcoplasm(GO:0016528) sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 1.2 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.8 | GO:0030141 | secretory granule(GO:0030141) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.5 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.3 | 1.3 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.2 | 1.6 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.2 | 1.0 | GO:0008469 | histone-arginine N-methyltransferase activity(GO:0008469) |
| 0.2 | 1.6 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.2 | 0.8 | GO:0019778 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.2 | 0.6 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.2 | 1.4 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.2 | 1.5 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.6 | GO:0051800 | inositol-1,3,4,5-tetrakisphosphate 3-phosphatase activity(GO:0051717) phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.1 | 0.4 | GO:0033857 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 0.3 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.1 | 0.8 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 0.5 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 0.5 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 0.6 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.1 | 1.9 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 1.5 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 1.3 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 1.6 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 2.5 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 0.8 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 1.5 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 0.3 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.8 | GO:0015149 | glucose transmembrane transporter activity(GO:0005355) monosaccharide transmembrane transporter activity(GO:0015145) hexose transmembrane transporter activity(GO:0015149) |
| 0.0 | 0.8 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.9 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 2.0 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.5 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 1.3 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.9 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 1.2 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.7 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 2.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 1.9 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.5 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.9 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.6 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.6 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.4 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.7 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.5 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 1.2 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.8 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 1.7 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.4 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 2.5 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 1.2 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 1.1 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.4 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 3.7 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 0.9 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 1.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.1 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 1.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.3 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.5 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.3 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.9 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.5 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.9 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.2 | 1.1 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.2 | 2.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.2 | 2.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 2.7 | REACTOME PROCESSIVE SYNTHESIS ON THE LAGGING STRAND | Genes involved in Processive synthesis on the lagging strand |
| 0.1 | 2.0 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 1.6 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 0.8 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 3.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 1.9 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 1.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.8 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |