Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
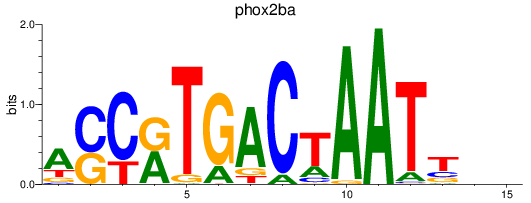
Results for phox2ba
Z-value: 0.84
Transcription factors associated with phox2ba
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
phox2ba
|
ENSDARG00000075330 | paired like homeobox 2Ba |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| phox2ba | dr11_v1_chr1_-_19079957_19079957 | -0.52 | 2.4e-02 | Click! |
Activity profile of phox2ba motif
Sorted Z-values of phox2ba motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_38161033 | 3.28 |
ENSDART00000145907
|
gucy2d
|
guanylate cyclase 2D, retinal |
| chr4_+_76509294 | 2.93 |
ENSDART00000099899
|
ms4a17a.17
|
membrane-spanning 4-domains, subfamily A, member 17A.17 |
| chr3_+_23247325 | 2.70 |
ENSDART00000114190
|
ppp1r9ba
|
protein phosphatase 1, regulatory subunit 9Ba |
| chr2_-_53481912 | 2.14 |
ENSDART00000189610
|
hsd11b1lb
|
hydroxysteroid (11-beta) dehydrogenase 1-like b |
| chr14_+_32839535 | 1.87 |
ENSDART00000168975
|
arr3b
|
arrestin 3b, retinal (X-arrestin) |
| chr14_-_45702721 | 1.79 |
ENSDART00000165727
|
si:dkey-226n24.1
|
si:dkey-226n24.1 |
| chr11_+_13223625 | 1.69 |
ENSDART00000161275
|
abcb11b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11b |
| chr4_+_7841627 | 1.62 |
ENSDART00000037997
|
ucmaa
|
upper zone of growth plate and cartilage matrix associated a |
| chr15_-_23908605 | 1.57 |
ENSDART00000185578
|
usp32
|
ubiquitin specific peptidase 32 |
| chr4_+_11375894 | 1.51 |
ENSDART00000190471
ENSDART00000143963 |
pcloa
|
piccolo presynaptic cytomatrix protein a |
| chr18_-_12612699 | 1.45 |
ENSDART00000090335
|
hipk2
|
homeodomain interacting protein kinase 2 |
| chr21_+_44300689 | 1.43 |
ENSDART00000186298
ENSDART00000142810 |
gabra3
|
gamma-aminobutyric acid (GABA) A receptor, alpha 3 |
| chr5_-_44829719 | 1.43 |
ENSDART00000019104
|
fbp2
|
fructose-1,6-bisphosphatase 2 |
| chr24_-_25574967 | 1.34 |
ENSDART00000189828
|
cnksr2a
|
connector enhancer of kinase suppressor of Ras 2a |
| chr19_-_10373630 | 1.32 |
ENSDART00000131517
|
si:ch211-232m10.6
|
si:ch211-232m10.6 |
| chr20_+_4060839 | 1.28 |
ENSDART00000178565
|
TRIM67
|
tripartite motif containing 67 |
| chr14_-_2355833 | 1.17 |
ENSDART00000157677
|
PCDHGC3 (1 of many)
|
si:ch73-233f7.6 |
| chr9_+_55321322 | 1.16 |
ENSDART00000111111
ENSDART00000191579 |
nlgn4b
|
neuroligin 4b |
| chr21_+_27189490 | 1.09 |
ENSDART00000125349
|
bada
|
BCL2 associated agonist of cell death a |
| chr13_-_34862452 | 1.07 |
ENSDART00000134573
ENSDART00000047552 |
sptlc3
|
serine palmitoyltransferase, long chain base subunit 3 |
| chr12_+_32368574 | 1.05 |
ENSDART00000086389
|
ANKFN1
|
si:ch211-277e21.2 |
| chr14_-_47314011 | 0.94 |
ENSDART00000178523
|
fstl5
|
follistatin-like 5 |
| chr16_+_47428721 | 0.89 |
ENSDART00000180597
ENSDART00000032309 |
slc25a32b
|
solute carrier family 25 (mitochondrial folate carrier), member 32b |
| chr17_+_27134806 | 0.83 |
ENSDART00000151901
|
rps6ka1
|
ribosomal protein S6 kinase a, polypeptide 1 |
| chr4_-_4834347 | 0.82 |
ENSDART00000141803
|
coa6
|
cytochrome c oxidase assembly factor 6 |
| chr15_+_2500510 | 0.82 |
ENSDART00000180329
|
wdr53
|
WD repeat domain 53 |
| chr2_-_51223658 | 0.76 |
ENSDART00000171770
|
pigrl4.2
|
polymeric immunoglobulin receptor-like 4.2 |
| chr21_+_40239192 | 0.74 |
ENSDART00000135857
|
or115-9
|
odorant receptor, family F, subfamily 115, member 9 |
| chr8_-_36370552 | 0.65 |
ENSDART00000097932
ENSDART00000148323 |
si:busm1-104n07.3
|
si:busm1-104n07.3 |
| chr11_-_23459779 | 0.60 |
ENSDART00000183935
ENSDART00000184125 ENSDART00000193284 |
plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr21_+_9689103 | 0.60 |
ENSDART00000165132
|
mapk10
|
mitogen-activated protein kinase 10 |
| chr5_-_25236340 | 0.59 |
ENSDART00000162774
|
abca2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr4_-_4834617 | 0.56 |
ENSDART00000141539
|
coa6
|
cytochrome c oxidase assembly factor 6 |
| chr4_-_77551860 | 0.56 |
ENSDART00000188176
|
AL935186.6
|
|
| chr7_+_72460911 | 0.54 |
ENSDART00000160682
ENSDART00000168532 |
HECTD4
|
HECT domain E3 ubiquitin protein ligase 4 |
| chr6_+_16493755 | 0.51 |
ENSDART00000126318
|
si:ch73-193c12.2
|
si:ch73-193c12.2 |
| chr19_-_32600138 | 0.47 |
ENSDART00000052101
|
zgc:91944
|
zgc:91944 |
| chr8_+_10862353 | 0.47 |
ENSDART00000140717
|
brpf3b
|
bromodomain and PHD finger containing, 3b |
| chr24_+_39227519 | 0.47 |
ENSDART00000184611
ENSDART00000193494 ENSDART00000190728 ENSDART00000168705 |
MPRIP
|
si:ch73-103b11.2 |
| chr20_-_18736281 | 0.45 |
ENSDART00000142837
|
enpp5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 |
| chr16_-_25519762 | 0.43 |
ENSDART00000146479
ENSDART00000142062 |
dtnbp1a
|
dystrobrevin binding protein 1a |
| chr8_+_10835456 | 0.42 |
ENSDART00000151388
|
mapk13
|
mitogen-activated protein kinase 13 |
| chr13_+_42858228 | 0.38 |
ENSDART00000179763
|
cdh23
|
cadherin-related 23 |
| chr9_-_54304684 | 0.32 |
ENSDART00000109512
|
il13
|
interleukin 13 |
| chr17_-_20717845 | 0.32 |
ENSDART00000150037
|
ank3b
|
ankyrin 3b |
| chr10_+_26645953 | 0.30 |
ENSDART00000131482
|
adgrg4b
|
adhesion G protein-coupled receptor G4b |
| chr18_+_27000850 | 0.29 |
ENSDART00000188906
ENSDART00000086131 |
pik3c2a
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 alpha |
| chr23_+_30048849 | 0.28 |
ENSDART00000126027
|
uts2a
|
urotensin 2, alpha |
| chr18_+_27598755 | 0.27 |
ENSDART00000193808
|
cd82b
|
CD82 molecule b |
| chr10_+_26646104 | 0.26 |
ENSDART00000187685
|
adgrg4b
|
adhesion G protein-coupled receptor G4b |
| chr5_-_44286987 | 0.25 |
ENSDART00000184112
|
si:ch73-337l15.2
|
si:ch73-337l15.2 |
| chr19_+_10799929 | 0.21 |
ENSDART00000159152
|
lipea
|
lipase, hormone-sensitive a |
| chr15_+_5360407 | 0.20 |
ENSDART00000110420
|
or112-1
|
odorant receptor, family A, subfamily 112, member 1 |
| chr1_-_33380011 | 0.20 |
ENSDART00000141347
ENSDART00000136383 |
cd99
|
CD99 molecule |
| chr7_-_30082931 | 0.20 |
ENSDART00000075600
|
tspan3b
|
tetraspanin 3b |
| chr18_-_5111449 | 0.20 |
ENSDART00000046902
|
pdcd10a
|
programmed cell death 10a |
| chr17_+_33719415 | 0.18 |
ENSDART00000132294
|
fut8a
|
fucosyltransferase 8a (alpha (1,6) fucosyltransferase) |
| chr8_-_11832771 | 0.18 |
ENSDART00000193412
ENSDART00000184720 ENSDART00000139947 |
rapgef1a
|
Rap guanine nucleotide exchange factor (GEF) 1a |
| chr19_-_32600638 | 0.17 |
ENSDART00000143497
|
zgc:91944
|
zgc:91944 |
| chr17_+_30587333 | 0.16 |
ENSDART00000156500
|
nhsl1a
|
NHS-like 1a |
| chr6_+_11850359 | 0.14 |
ENSDART00000109552
ENSDART00000188139 ENSDART00000181499 ENSDART00000178269 |
baz2ba
|
bromodomain adjacent to zinc finger domain, 2Ba |
| chr25_+_32530976 | 0.13 |
ENSDART00000156190
ENSDART00000103324 |
scaper
|
S-phase cyclin A-associated protein in the ER |
| chr14_+_45702521 | 0.13 |
ENSDART00000112470
|
ccdc88b
|
coiled-coil domain containing 88B |
| chr15_+_25681044 | 0.12 |
ENSDART00000077853
|
hic1
|
hypermethylated in cancer 1 |
| chr21_+_40225915 | 0.11 |
ENSDART00000048475
ENSDART00000174122 |
or115-12
|
odorant receptor, family F, subfamily 115, member 12 |
| chr8_+_31821396 | 0.10 |
ENSDART00000077053
|
plcxd3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr24_-_27436319 | 0.10 |
ENSDART00000171489
|
si:dkey-25o1.7
|
si:dkey-25o1.7 |
| chr2_+_55982940 | 0.08 |
ENSDART00000097753
ENSDART00000097751 |
nmrk2
|
nicotinamide riboside kinase 2 |
| chr5_-_44286778 | 0.06 |
ENSDART00000146080
|
si:ch73-337l15.2
|
si:ch73-337l15.2 |
| chr3_+_57788761 | 0.05 |
ENSDART00000149741
|
si:ch73-362i18.1
|
si:ch73-362i18.1 |
| chr8_-_11835607 | 0.04 |
ENSDART00000183601
|
rapgef1a
|
Rap guanine nucleotide exchange factor (GEF) 1a |
Network of associatons between targets according to the STRING database.
First level regulatory network of phox2ba
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0005985 | sucrose metabolic process(GO:0005985) sucrose biosynthetic process(GO:0005986) disaccharide biosynthetic process(GO:0046351) |
| 0.4 | 1.7 | GO:0032782 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.2 | 1.5 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) |
| 0.2 | 1.9 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.2 | 1.5 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.1 | 3.3 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 1.6 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) |
| 0.1 | 0.4 | GO:0060155 | secretory granule organization(GO:0033363) platelet dense granule organization(GO:0060155) |
| 0.1 | 1.1 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.1 | 0.4 | GO:0035678 | neuromast hair cell morphogenesis(GO:0035678) |
| 0.1 | 1.4 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.1 | 0.2 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 0.1 | 1.4 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.0 | 0.2 | GO:0072672 | neutrophil extravasation(GO:0072672) regulation of neutrophil extravasation(GO:2000389) positive regulation of neutrophil extravasation(GO:2000391) |
| 0.0 | 0.5 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 2.7 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.2 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.2 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.2 | GO:0036065 | fucosylation(GO:0036065) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.1 | 1.1 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 0.2 | GO:0090443 | FAR/SIN/STRIPAK complex(GO:0090443) |
| 0.0 | 1.4 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 1.5 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.4 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.5 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 2.7 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 0.4 | GO:0032420 | stereocilium(GO:0032420) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.4 | 1.7 | GO:0015432 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.3 | 0.9 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.2 | 3.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.2 | 1.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 1.5 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 1.4 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 1.5 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.1 | 2.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.8 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 0.6 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.6 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.5 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.2 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.0 | 0.2 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.3 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.4 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.8 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.6 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 1.5 | PID P53 REGULATION PATHWAY | p53 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 0.6 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 0.8 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.1 | 0.4 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 1.4 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.6 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |