Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
Results for pgr
Z-value: 0.69
Transcription factors associated with pgr
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pgr
|
ENSDARG00000035966 | progesterone receptor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pgr | dr11_v1_chr18_+_41820039_41820039 | 0.37 | 1.2e-01 | Click! |
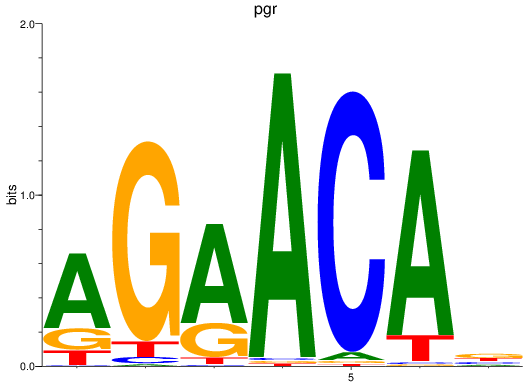
Activity profile of pgr motif
Sorted Z-values of pgr motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_59348118 | 3.04 |
ENSDART00000170901
|
cyp3a65
|
cytochrome P450, family 3, subfamily A, polypeptide 65 |
| chr9_-_22272181 | 2.23 |
ENSDART00000113174
|
crygm2d7
|
crystallin, gamma M2d7 |
| chr21_+_25187210 | 2.16 |
ENSDART00000101147
ENSDART00000167528 |
si:dkey-183i3.5
|
si:dkey-183i3.5 |
| chr1_+_51615672 | 1.91 |
ENSDART00000165117
|
zgc:165656
|
zgc:165656 |
| chr17_-_6730247 | 1.90 |
ENSDART00000031091
|
vsnl1b
|
visinin-like 1b |
| chr1_+_7538909 | 1.88 |
ENSDART00000148276
|
mylz3
|
myosin, light polypeptide 3, skeletal muscle |
| chr3_+_16612574 | 1.81 |
ENSDART00000104481
|
slc17a7a
|
solute carrier family 17 (vesicular glutamate transporter), member 7a |
| chr24_-_27473771 | 1.78 |
ENSDART00000139874
|
cxl34b.11
|
CX chemokine ligand 34b, duplicate 11 |
| chr5_+_45677781 | 1.72 |
ENSDART00000163120
ENSDART00000126537 |
gc
|
group-specific component (vitamin D binding protein) |
| chr9_-_43001898 | 1.70 |
ENSDART00000138515
|
ttn.2
|
titin, tandem duplicate 2 |
| chr18_+_38908903 | 1.61 |
ENSDART00000159834
|
myo5aa
|
myosin VAa |
| chr19_-_29437108 | 1.43 |
ENSDART00000052108
ENSDART00000074497 |
fndc5b
|
fibronectin type III domain containing 5b |
| chr12_-_3237561 | 1.39 |
ENSDART00000164665
|
si:ch1073-13h15.3
|
si:ch1073-13h15.3 |
| chr22_-_278328 | 1.35 |
ENSDART00000098072
|
myhz1.1
|
myosin, heavy polypeptide 1.1, skeletal muscle |
| chr7_+_39389273 | 1.35 |
ENSDART00000191298
|
tnni2b.2
|
troponin I type 2b (skeletal, fast), tandem duplicate 2 |
| chr3_-_28048475 | 1.34 |
ENSDART00000150888
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr7_+_2467057 | 1.34 |
ENSDART00000154517
|
si:dkey-125e8.3
|
si:dkey-125e8.3 |
| chr2_+_10134345 | 1.28 |
ENSDART00000100725
|
ahsg2
|
alpha-2-HS-glycoprotein 2 |
| chr20_-_8419057 | 1.26 |
ENSDART00000145841
|
dab1a
|
Dab, reelin signal transducer, homolog 1a (Drosophila) |
| chr20_+_30445971 | 1.22 |
ENSDART00000153150
|
myt1la
|
myelin transcription factor 1-like, a |
| chr24_+_3307857 | 1.21 |
ENSDART00000106527
|
gyg1b
|
glycogenin 1b |
| chr6_+_41096058 | 1.20 |
ENSDART00000028373
|
fkbp5
|
FK506 binding protein 5 |
| chr3_+_34220194 | 1.20 |
ENSDART00000145859
|
slc25a23b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23b |
| chr21_+_25643880 | 1.20 |
ENSDART00000192392
ENSDART00000145091 |
tmem151a
|
transmembrane protein 151A |
| chr2_+_47582488 | 1.15 |
ENSDART00000149967
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr11_-_1024483 | 1.15 |
ENSDART00000017551
|
slc6a1b
|
solute carrier family 6 (neurotransmitter transporter), member 1b |
| chr5_+_13472234 | 1.15 |
ENSDART00000114069
ENSDART00000132406 |
cnnm4b
|
cyclin and CBS domain divalent metal cation transport mediator 4b |
| chr10_-_22249444 | 1.11 |
ENSDART00000148831
|
fgf11b
|
fibroblast growth factor 11b |
| chr12_-_4028079 | 1.10 |
ENSDART00000128676
|
si:ch211-180a12.2
|
si:ch211-180a12.2 |
| chr21_-_10446405 | 1.10 |
ENSDART00000167948
|
hcn1
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 1 |
| chr10_+_21786656 | 1.10 |
ENSDART00000185851
ENSDART00000167219 |
pcdh1g26
|
protocadherin 1 gamma 26 |
| chr1_+_51896147 | 1.10 |
ENSDART00000167139
|
nfixa
|
nuclear factor I/Xa |
| chr11_+_13223625 | 1.10 |
ENSDART00000161275
|
abcb11b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11b |
| chr2_+_52232630 | 1.09 |
ENSDART00000006216
|
plpp2a
|
phospholipid phosphatase 2a |
| chr5_+_51102010 | 1.05 |
ENSDART00000110377
|
zgc:194398
|
zgc:194398 |
| chr10_+_375042 | 1.04 |
ENSDART00000171854
|
si:ch1073-303d10.1
|
si:ch1073-303d10.1 |
| chr11_-_33919014 | 1.04 |
ENSDART00000181203
|
phka2
|
phosphorylase kinase, alpha 2 (liver) |
| chr7_-_33868903 | 1.03 |
ENSDART00000173500
ENSDART00000178746 |
uacab
|
uveal autoantigen with coiled-coil domains and ankyrin repeats b |
| chr19_+_9344171 | 1.03 |
ENSDART00000133447
ENSDART00000104622 |
si:ch211-288g17.4
|
si:ch211-288g17.4 |
| chr5_+_30680804 | 1.03 |
ENSDART00000078049
|
pdzd3b
|
PDZ domain containing 3b |
| chr21_-_26089964 | 1.01 |
ENSDART00000027848
|
tlcd1
|
TLC domain containing 1 |
| chr6_-_27139396 | 1.01 |
ENSDART00000055848
|
zgc:103559
|
zgc:103559 |
| chr8_-_14050758 | 0.99 |
ENSDART00000133922
|
atp2b3a
|
ATPase plasma membrane Ca2+ transporting 3a |
| chr18_+_8805575 | 0.99 |
ENSDART00000137098
|
impdh1a
|
IMP (inosine 5'-monophosphate) dehydrogenase 1a |
| chr25_-_210730 | 0.98 |
ENSDART00000187580
|
FP236318.1
|
|
| chr8_-_10045043 | 0.94 |
ENSDART00000081917
|
si:dkey-8e10.3
|
si:dkey-8e10.3 |
| chr23_-_45504991 | 0.94 |
ENSDART00000148761
|
col24a1
|
collagen type XXIV alpha 1 |
| chr5_+_62356304 | 0.94 |
ENSDART00000148381
|
aspa
|
aspartoacylase |
| chr25_+_31277415 | 0.92 |
ENSDART00000036275
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr8_+_30780985 | 0.90 |
ENSDART00000193971
ENSDART00000111533 |
tas2r200.2
|
taste receptor, type 2, member 200, tandem duplicate 2 |
| chr3_+_16449490 | 0.90 |
ENSDART00000141467
|
kcnj12b
|
potassium inwardly-rectifying channel, subfamily J, member 12b |
| chr7_-_71486162 | 0.90 |
ENSDART00000045253
|
aco1
|
aconitase 1, soluble |
| chr3_-_11922713 | 0.90 |
ENSDART00000150575
|
coro7
|
coronin 7 |
| chr15_+_32710300 | 0.89 |
ENSDART00000167857
|
postnb
|
periostin, osteoblast specific factor b |
| chr9_-_41277347 | 0.88 |
ENSDART00000181213
|
glsb
|
glutaminase b |
| chr5_-_25174420 | 0.88 |
ENSDART00000141554
|
abca2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr11_+_13224281 | 0.87 |
ENSDART00000102557
ENSDART00000178706 |
abcb11b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11b |
| chr1_+_56229674 | 0.86 |
ENSDART00000127714
|
c3a.2
|
complement component c3a, duplicate 2 |
| chr8_+_53452681 | 0.86 |
ENSDART00000166705
|
cacna1db
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, b |
| chr6_+_11250033 | 0.85 |
ENSDART00000065411
ENSDART00000132677 |
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr15_+_12030367 | 0.84 |
ENSDART00000176178
|
FO904901.1
|
|
| chr17_+_36627099 | 0.84 |
ENSDART00000154104
|
impg1b
|
interphotoreceptor matrix proteoglycan 1b |
| chr18_-_46063773 | 0.83 |
ENSDART00000078561
|
si:ch73-262h23.4
|
si:ch73-262h23.4 |
| chr20_+_18580176 | 0.83 |
ENSDART00000185310
|
si:dkeyp-72h1.1
|
si:dkeyp-72h1.1 |
| chr14_-_2187202 | 0.83 |
ENSDART00000160205
|
pcdh2ab12
|
protocadherin 2 alpha b 12 |
| chr6_-_1865323 | 0.83 |
ENSDART00000155775
|
dlgap4b
|
discs, large (Drosophila) homolog-associated protein 4b |
| chr11_+_21910343 | 0.82 |
ENSDART00000161485
|
foxp4
|
forkhead box P4 |
| chr2_+_47581997 | 0.82 |
ENSDART00000112579
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr11_+_21076872 | 0.81 |
ENSDART00000155521
|
prelp
|
proline/arginine-rich end leucine-rich repeat protein |
| chr5_-_32308526 | 0.81 |
ENSDART00000193758
|
myhz1.1
|
myosin, heavy polypeptide 1.1, skeletal muscle |
| chr7_+_73780936 | 0.80 |
ENSDART00000092691
|
pea15
|
proliferation and apoptosis adaptor protein 15 |
| chr15_+_37197494 | 0.80 |
ENSDART00000166203
|
aplp1
|
amyloid beta (A4) precursor-like protein 1 |
| chr13_-_44285793 | 0.79 |
ENSDART00000167383
|
CABZ01069436.1
|
|
| chr14_-_2209742 | 0.78 |
ENSDART00000054889
|
pcdh2ab5
|
protocadherin 2 alpha b 5 |
| chr15_+_47386939 | 0.77 |
ENSDART00000128224
|
FO904873.1
|
|
| chr7_-_2163361 | 0.76 |
ENSDART00000173654
|
si:cabz01007812.1
|
si:cabz01007812.1 |
| chr13_-_23264724 | 0.74 |
ENSDART00000051886
|
si:dkey-103j14.5
|
si:dkey-103j14.5 |
| chr20_+_43942278 | 0.73 |
ENSDART00000100571
|
clic5b
|
chloride intracellular channel 5b |
| chr5_-_37116265 | 0.73 |
ENSDART00000057613
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr15_+_28482862 | 0.73 |
ENSDART00000015286
ENSDART00000154320 |
ankrd13b
|
ankyrin repeat domain 13B |
| chr4_+_74551606 | 0.72 |
ENSDART00000174339
|
kcna1b
|
potassium voltage-gated channel, shaker-related subfamily, member 1b |
| chr15_-_24869826 | 0.72 |
ENSDART00000127047
|
tusc5a
|
tumor suppressor candidate 5a |
| chr15_-_163586 | 0.72 |
ENSDART00000163597
|
SEPT4
|
septin-4 |
| chr25_-_19090479 | 0.71 |
ENSDART00000027465
ENSDART00000177670 |
cacna2d4b
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4b |
| chr10_+_44146287 | 0.71 |
ENSDART00000162176
|
grk3
|
G protein-coupled receptor kinase 3 |
| chr8_+_68864 | 0.71 |
ENSDART00000164574
|
prr16
|
proline rich 16 |
| chr2_-_3678029 | 0.70 |
ENSDART00000146861
|
mmp16b
|
matrix metallopeptidase 16b (membrane-inserted) |
| chr6_-_60031693 | 0.70 |
ENSDART00000160275
|
CABZ01079262.1
|
|
| chr7_+_25033924 | 0.70 |
ENSDART00000170873
|
sb:cb1058
|
sb:cb1058 |
| chr9_-_22831836 | 0.70 |
ENSDART00000142585
|
neb
|
nebulin |
| chr5_+_11812089 | 0.69 |
ENSDART00000111359
|
fbxo21
|
F-box protein 21 |
| chr12_+_23875318 | 0.68 |
ENSDART00000152869
|
svila
|
supervillin a |
| chr14_-_2050057 | 0.66 |
ENSDART00000112875
|
PCDHB15
|
protocadherin beta 15 |
| chr14_-_2193248 | 0.65 |
ENSDART00000128659
|
pcdh2ab10
|
protocadherin 2 alpha b 10 |
| chr20_+_33532296 | 0.65 |
ENSDART00000153153
|
kcnf1a
|
potassium voltage-gated channel, subfamily F, member 1a |
| chr2_+_47582681 | 0.65 |
ENSDART00000187579
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr1_-_22687913 | 0.65 |
ENSDART00000168171
|
fgfbp2b
|
fibroblast growth factor binding protein 2b |
| chr5_+_69981453 | 0.65 |
ENSDART00000143860
|
si:ch211-154e10.1
|
si:ch211-154e10.1 |
| chr15_+_44472356 | 0.65 |
ENSDART00000155763
ENSDART00000028011 |
msantd4
|
Myb/SANT-like DNA-binding domain containing 4 with coiled-coils |
| chr2_-_24996441 | 0.64 |
ENSDART00000144795
|
slc35g2a
|
solute carrier family 35, member G2a |
| chr20_+_29587995 | 0.64 |
ENSDART00000153339
|
adam17b
|
ADAM metallopeptidase domain 17b |
| chr25_+_4541211 | 0.64 |
ENSDART00000129978
|
pnpla2
|
patatin-like phospholipase domain containing 2 |
| chr20_+_23185530 | 0.63 |
ENSDART00000142346
|
lrrc66
|
leucine rich repeat containing 66 |
| chr1_-_51615437 | 0.63 |
ENSDART00000152185
ENSDART00000152237 ENSDART00000129052 ENSDART00000152595 |
FBXO48
|
si:dkey-202b17.4 |
| chr20_-_35706379 | 0.63 |
ENSDART00000152847
|
si:dkey-30j22.1
|
si:dkey-30j22.1 |
| chr10_-_28519505 | 0.63 |
ENSDART00000137964
|
bbx
|
bobby sox homolog (Drosophila) |
| chr5_-_28029558 | 0.61 |
ENSDART00000078649
|
abcb9
|
ATP-binding cassette, sub-family B (MDR/TAP), member 9 |
| chr20_+_16750177 | 0.61 |
ENSDART00000185357
|
calm1b
|
calmodulin 1b |
| chr6_-_20707846 | 0.61 |
ENSDART00000169836
|
col18a1b
|
collagen type XVIII alpha 1 chain b |
| chr7_+_25036188 | 0.61 |
ENSDART00000163957
ENSDART00000169749 |
sb:cb1058
|
sb:cb1058 |
| chr1_+_51615506 | 0.61 |
ENSDART00000152767
|
zgc:165656
|
zgc:165656 |
| chr2_+_55916911 | 0.60 |
ENSDART00000189483
ENSDART00000183647 ENSDART00000083470 |
atcayb
|
ataxia, cerebellar, Cayman type b |
| chr18_-_10995410 | 0.60 |
ENSDART00000136751
|
tspan33b
|
tetraspanin 33b |
| chr8_+_9715010 | 0.60 |
ENSDART00000139414
|
gripap1
|
GRIP1 associated protein 1 |
| chr16_-_36099492 | 0.59 |
ENSDART00000180905
|
CU499336.2
|
|
| chr2_+_44571200 | 0.58 |
ENSDART00000098132
|
klhl24a
|
kelch-like family member 24a |
| chr21_-_14803554 | 0.58 |
ENSDART00000121853
|
si:dkey-11o18.5
|
si:dkey-11o18.5 |
| chr18_-_16179129 | 0.58 |
ENSDART00000125353
|
slc6a15
|
solute carrier family 6 (neutral amino acid transporter), member 15 |
| chr9_+_31752628 | 0.58 |
ENSDART00000060054
|
itgbl1
|
integrin, beta-like 1 |
| chr2_-_36932253 | 0.58 |
ENSDART00000124217
|
map1sb
|
microtubule-associated protein 1Sb |
| chr13_-_25719628 | 0.57 |
ENSDART00000135383
|
si:dkey-192p21.6
|
si:dkey-192p21.6 |
| chr6_-_40429411 | 0.57 |
ENSDART00000156005
ENSDART00000156357 |
si:dkey-28n18.9
|
si:dkey-28n18.9 |
| chr24_+_32592748 | 0.57 |
ENSDART00000188256
|
si:ch211-282b22.1
|
si:ch211-282b22.1 |
| chr7_-_66864756 | 0.57 |
ENSDART00000184462
ENSDART00000189424 |
ampd3a
|
adenosine monophosphate deaminase 3a |
| chr1_+_33969015 | 0.57 |
ENSDART00000042984
ENSDART00000146530 |
epha6
|
eph receptor A6 |
| chr18_-_341489 | 0.57 |
ENSDART00000168333
|
si:ch1073-83b15.1
|
si:ch1073-83b15.1 |
| chr2_-_24603325 | 0.56 |
ENSDART00000113356
|
crtc1a
|
CREB regulated transcription coactivator 1a |
| chr9_-_42871756 | 0.56 |
ENSDART00000191396
|
ttn.1
|
titin, tandem duplicate 1 |
| chr10_+_29199172 | 0.56 |
ENSDART00000148828
|
picalma
|
phosphatidylinositol binding clathrin assembly protein a |
| chr22_-_968484 | 0.56 |
ENSDART00000105895
|
cacna1sa
|
calcium channel, voltage-dependent, L type, alpha 1S subunit, a |
| chr7_+_61764040 | 0.56 |
ENSDART00000056745
|
acox3
|
acyl-CoA oxidase 3, pristanoyl |
| chr15_+_32710094 | 0.56 |
ENSDART00000159592
|
postnb
|
periostin, osteoblast specific factor b |
| chr6_+_11249706 | 0.55 |
ENSDART00000186547
ENSDART00000193287 |
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr7_-_51953613 | 0.54 |
ENSDART00000142042
|
cyp2x10.2
|
cytochrome P450, family 2, subfamily X, polypeptide 10.2 |
| chr15_+_15316326 | 0.54 |
ENSDART00000125864
|
camkk1b
|
calcium/calmodulin-dependent protein kinase kinase 1, alpha b |
| chr9_+_34127005 | 0.54 |
ENSDART00000167384
ENSDART00000078065 |
f5
|
coagulation factor V |
| chr11_+_35364445 | 0.54 |
ENSDART00000125766
|
camkvb
|
CaM kinase-like vesicle-associated b |
| chr10_+_21722892 | 0.53 |
ENSDART00000162855
|
pcdh1g13
|
protocadherin 1 gamma 13 |
| chr7_+_56253914 | 0.53 |
ENSDART00000073594
|
ankrd11
|
ankyrin repeat domain 11 |
| chr8_-_42677086 | 0.53 |
ENSDART00000191173
|
si:ch73-138n13.1
|
si:ch73-138n13.1 |
| chr11_-_28911172 | 0.53 |
ENSDART00000168493
|
igsf21a
|
immunoglobin superfamily, member 21a |
| chr20_-_27857676 | 0.53 |
ENSDART00000192934
|
syndig1l
|
synapse differentiation inducing 1-like |
| chr6_+_40874206 | 0.52 |
ENSDART00000004295
|
zgc:112163
|
zgc:112163 |
| chr25_-_207214 | 0.52 |
ENSDART00000193448
|
FP236318.3
|
|
| chr19_-_9867001 | 0.52 |
ENSDART00000091695
|
cacng7a
|
calcium channel, voltage-dependent, gamma subunit 7a |
| chr7_+_30823749 | 0.52 |
ENSDART00000085661
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr7_+_27977065 | 0.51 |
ENSDART00000089574
|
tub
|
tubby bipartite transcription factor |
| chr3_+_49397115 | 0.51 |
ENSDART00000176042
|
tecra
|
trans-2,3-enoyl-CoA reductase a |
| chr5_-_71460556 | 0.51 |
ENSDART00000108804
|
brinp1
|
bone morphogenetic protein/retinoic acid inducible neural-specific 1 |
| chr6_+_41099787 | 0.51 |
ENSDART00000186884
|
fkbp5
|
FK506 binding protein 5 |
| chr18_+_1145571 | 0.51 |
ENSDART00000135055
|
rec114
|
REC114 meiotic recombination protein |
| chr17_-_465285 | 0.51 |
ENSDART00000168718
|
chrm5a
|
cholinergic receptor, muscarinic 5a |
| chr21_+_18353703 | 0.50 |
ENSDART00000181396
ENSDART00000166359 |
si:ch73-287m6.1
|
si:ch73-287m6.1 |
| chr6_-_18400548 | 0.49 |
ENSDART00000179797
ENSDART00000164891 |
trim25
|
tripartite motif containing 25 |
| chr4_-_14328997 | 0.49 |
ENSDART00000091151
|
nell2b
|
neural EGFL like 2b |
| chr17_-_26507289 | 0.49 |
ENSDART00000155616
|
ccser2a
|
coiled-coil serine-rich protein 2a |
| chr8_-_36475328 | 0.49 |
ENSDART00000048448
|
si:busm1-266f07.2
|
si:busm1-266f07.2 |
| chr16_+_16933002 | 0.48 |
ENSDART00000173163
|
myh14
|
myosin, heavy chain 14, non-muscle |
| chr2_-_55337585 | 0.48 |
ENSDART00000177924
|
tpm4b
|
tropomyosin 4b |
| chr4_-_16333944 | 0.47 |
ENSDART00000079523
|
epyc
|
epiphycan |
| chr1_+_58456494 | 0.47 |
ENSDART00000164130
|
si:ch73-236c18.5
|
si:ch73-236c18.5 |
| chr3_+_22935183 | 0.47 |
ENSDART00000157378
|
hdac5
|
histone deacetylase 5 |
| chr4_-_20181964 | 0.47 |
ENSDART00000022539
|
fgl2a
|
fibrinogen-like 2a |
| chr21_+_25221940 | 0.47 |
ENSDART00000108972
|
sycn.1
|
syncollin, tandem duplicate 1 |
| chr21_-_22737228 | 0.46 |
ENSDART00000151366
|
fbxo40.2
|
F-box protein 40, tandem duplicate 2 |
| chr13_+_38430466 | 0.46 |
ENSDART00000132691
|
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr4_+_3980247 | 0.46 |
ENSDART00000049194
|
gpr37b
|
G protein-coupled receptor 37b |
| chr12_-_41684729 | 0.46 |
ENSDART00000184461
|
jakmip3
|
Janus kinase and microtubule interacting protein 3 |
| chr1_-_17803614 | 0.46 |
ENSDART00000138475
|
sorbs2a
|
sorbin and SH3 domain containing 2a |
| chr17_+_25677458 | 0.45 |
ENSDART00000029703
|
kcnh1a
|
potassium voltage-gated channel, subfamily H (eag-related), member 1a |
| chr8_+_36803415 | 0.45 |
ENSDART00000111680
|
iqsec2b
|
IQ motif and Sec7 domain 2b |
| chr1_-_65017 | 0.45 |
ENSDART00000168609
|
si:zfos-1011f11.2
|
si:zfos-1011f11.2 |
| chr7_+_17229980 | 0.45 |
ENSDART00000184910
|
slc6a5
|
solute carrier family 6 (neurotransmitter transporter), member 5 |
| chr5_+_52706316 | 0.44 |
ENSDART00000169488
|
apba1a
|
amyloid beta (A4) precursor protein-binding, family A, member 1a |
| chr3_-_42981739 | 0.44 |
ENSDART00000167844
|
mafk
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog K |
| chr6_-_49078263 | 0.44 |
ENSDART00000032982
|
slc5a8l
|
solute carrier family 5 (iodide transporter), member 8-like |
| chr13_-_50309969 | 0.44 |
ENSDART00000191597
|
CU928131.1
|
|
| chr7_-_52842007 | 0.44 |
ENSDART00000182710
|
map1aa
|
microtubule-associated protein 1Aa |
| chr8_-_12468744 | 0.44 |
ENSDART00000135019
|
FIBCD1 (1 of many)
|
si:dkeyp-51b7.3 |
| chr24_-_22702017 | 0.44 |
ENSDART00000179403
|
ctnnd2a
|
catenin (cadherin-associated protein), delta 2a |
| chr11_-_43948885 | 0.43 |
ENSDART00000164522
|
CABZ01074203.1
|
|
| chr4_+_70967639 | 0.43 |
ENSDART00000159230
|
si:dkeyp-80d11.12
|
si:dkeyp-80d11.12 |
| chr9_+_4306122 | 0.43 |
ENSDART00000193722
ENSDART00000190521 |
kalrna
|
kalirin RhoGEF kinase a |
| chr17_-_39886628 | 0.43 |
ENSDART00000002217
|
zmp:0000000545
|
zmp:0000000545 |
| chr25_-_12203952 | 0.43 |
ENSDART00000158204
ENSDART00000091727 |
ntrk3a
|
neurotrophic tyrosine kinase, receptor, type 3a |
| chr4_+_26628822 | 0.42 |
ENSDART00000191030
ENSDART00000186113 ENSDART00000186764 ENSDART00000165158 |
iqsec3a
|
IQ motif and Sec7 domain 3a |
| chr6_+_515181 | 0.42 |
ENSDART00000171374
|
CACNA1I (1 of many)
|
si:ch73-379f7.5 |
| chr22_-_20575679 | 0.42 |
ENSDART00000089033
|
lingo3a
|
leucine rich repeat and Ig domain containing 3a |
| chr7_-_44672095 | 0.42 |
ENSDART00000073745
|
cmtm4
|
CKLF-like MARVEL transmembrane domain containing 4 |
| chr7_-_74090168 | 0.42 |
ENSDART00000050528
|
tyrp1a
|
tyrosinase-related protein 1a |
| chr19_-_42651615 | 0.42 |
ENSDART00000123360
|
susd5
|
sushi domain containing 5 |
| chr2_+_5300405 | 0.42 |
ENSDART00000019925
|
GNB4
|
G protein subunit beta 4 |
| chr6_-_8489810 | 0.42 |
ENSDART00000124643
|
rasal3
|
RAS protein activator like 3 |
| chr17_-_8169774 | 0.41 |
ENSDART00000091828
|
syne1b
|
spectrin repeat containing, nuclear envelope 1b |
| chr16_-_6821927 | 0.41 |
ENSDART00000149070
ENSDART00000149570 |
mbpb
|
myelin basic protein b |
| chr6_+_23887314 | 0.41 |
ENSDART00000163188
|
znf648
|
zinc finger protein 648 |
| chr1_+_38858399 | 0.41 |
ENSDART00000165454
|
CU915762.1
|
|
| chr18_+_27101096 | 0.40 |
ENSDART00000171128
ENSDART00000086094 |
plekha7a
|
pleckstrin homology domain containing, family A member 7a |
| chr7_+_33279108 | 0.39 |
ENSDART00000084530
|
coro2ba
|
coronin, actin binding protein, 2Ba |
| chr16_-_2414063 | 0.39 |
ENSDART00000073621
|
zgc:152945
|
zgc:152945 |
Network of associatons between targets according to the STRING database.
First level regulatory network of pgr
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0015722 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.3 | 1.0 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.3 | 1.0 | GO:0006145 | allantoin catabolic process(GO:0000256) purine nucleobase catabolic process(GO:0006145) |
| 0.3 | 1.0 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.2 | 1.8 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.2 | 0.9 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.2 | 1.1 | GO:0038065 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.2 | 0.7 | GO:0006530 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.2 | 0.5 | GO:0039535 | RIG-I signaling pathway(GO:0039529) regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039531) regulation of RIG-I signaling pathway(GO:0039535) |
| 0.2 | 0.6 | GO:0010898 | regulation of triglyceride catabolic process(GO:0010896) positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.2 | 1.3 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.1 | 0.6 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.1 | 0.6 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 | 2.3 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.1 | 0.6 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 | 1.4 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.1 | 0.4 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 0.7 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.1 | 1.2 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.1 | 0.9 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.1 | 1.1 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.1 | 1.2 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 0.5 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.1 | 0.6 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.1 | 0.6 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 1.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.4 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.1 | 0.3 | GO:0045938 | positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.1 | 0.8 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.1 | 0.3 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 1.2 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.1 | 0.7 | GO:0003315 | heart rudiment formation(GO:0003315) |
| 0.1 | 2.6 | GO:1903670 | regulation of sprouting angiogenesis(GO:1903670) |
| 0.1 | 0.3 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.1 | 1.7 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.1 | 0.2 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) Schwann cell migration(GO:0036135) cardiac neuron differentiation(GO:0060945) cardiac neuron development(GO:0060959) |
| 0.1 | 0.6 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 1.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 1.1 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 0.5 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.3 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 1.3 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.8 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 0.4 | GO:0042438 | melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) |
| 0.0 | 0.5 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.7 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 0.3 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.2 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 2.2 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 1.0 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.2 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.4 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 1.0 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.5 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.3 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.4 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.0 | 0.2 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.1 | GO:0034773 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.3 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.2 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 3.9 | GO:0009410 | response to xenobiotic stimulus(GO:0009410) |
| 0.0 | 0.4 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.3 | GO:0006007 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.4 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.0 | 1.7 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.5 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.0 | 0.2 | GO:0060005 | reflex(GO:0060004) vestibular reflex(GO:0060005) |
| 0.0 | 1.3 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 0.1 | GO:0048521 | negative regulation of behavior(GO:0048521) negative regulation of feeding behavior(GO:2000252) |
| 0.0 | 0.3 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.1 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.0 | 0.1 | GO:0045852 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.0 | 0.8 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.5 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.3 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.0 | 0.7 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 0.4 | GO:1990090 | response to nerve growth factor(GO:1990089) cellular response to nerve growth factor stimulus(GO:1990090) |
| 0.0 | 0.7 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.3 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.2 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.6 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.9 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.3 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.8 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 0.9 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 5.5 | GO:0034765 | regulation of ion transmembrane transport(GO:0034765) |
| 0.0 | 0.1 | GO:2001287 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.4 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.0 | 0.1 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.0 | 0.8 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.4 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 3.1 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.5 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.4 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.5 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.7 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 3.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.7 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.2 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.0 | 0.1 | GO:0070293 | renal absorption(GO:0070293) |
| 0.0 | 1.1 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.5 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.0 | 1.1 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.9 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 1.4 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 0.2 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.3 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.3 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) synaptic vesicle budding(GO:0070142) |
| 0.0 | 0.2 | GO:0070570 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 0.3 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.4 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 0.1 | GO:0016119 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.1 | 0.6 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.1 | 0.6 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.1 | 1.8 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 0.8 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.1 | 2.5 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 0.5 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 0.4 | GO:0005913 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.1 | 1.1 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.7 | GO:0033270 | paranode region of axon(GO:0033270) juxtaparanode region of axon(GO:0044224) |
| 0.1 | 0.6 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 0.6 | GO:0098894 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.1 | 2.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 1.4 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 2.1 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.3 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.0 | 4.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.4 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 2.9 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.3 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.0 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.4 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 2.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.3 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 1.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 1.5 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.3 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 1.5 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 4.4 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.6 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.8 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.2 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.5 | 2.0 | GO:0015126 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.4 | 1.7 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.3 | 1.0 | GO:0010852 | cyclase inhibitor activity(GO:0010852) guanylate cyclase inhibitor activity(GO:0030251) |
| 0.3 | 0.9 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 0.3 | 1.0 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.2 | 0.6 | GO:0046978 | TAP1 binding(GO:0046978) |
| 0.2 | 2.5 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.1 | 0.7 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.1 | 1.8 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.1 | 0.4 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.6 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.1 | 0.3 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.6 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.1 | 0.4 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.1 | 0.9 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.3 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 1.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 0.6 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 1.3 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 0.4 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.1 | 0.4 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 0.8 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.1 | 1.1 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 1.7 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.2 | GO:0008887 | glycerate kinase activity(GO:0008887) |
| 0.1 | 0.3 | GO:0031841 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.1 | 1.1 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 1.6 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.9 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 0.4 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.5 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.3 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.4 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 1.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.4 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.1 | GO:0043121 | neurotrophin binding(GO:0043121) |
| 0.0 | 0.5 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.9 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.7 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.2 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 3.5 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 1.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 1.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 1.0 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.1 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.3 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.6 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.4 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 1.3 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.3 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 1.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 2.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.0 | 1.1 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.5 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 1.6 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.2 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 0.1 | GO:0072571 | ADP-D-ribose binding(GO:0072570) mono-ADP-D-ribose binding(GO:0072571) |
| 0.0 | 0.3 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.6 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.3 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 3.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 2.5 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.3 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.2 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 4.1 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.2 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.3 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.8 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.8 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.2 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.7 | GO:0005244 | voltage-gated ion channel activity(GO:0005244) |
| 0.0 | 1.2 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.6 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.1 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 1.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.0 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 1.0 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.1 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.4 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 1.7 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.6 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.3 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.4 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.5 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.1 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.8 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.2 | PID IL2 1PATHWAY | IL2-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 0.9 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 1.0 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 1.6 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.6 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.5 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.4 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.0 | 0.6 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.4 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.1 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.3 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |