Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
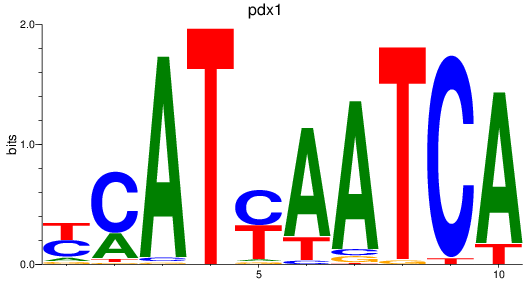
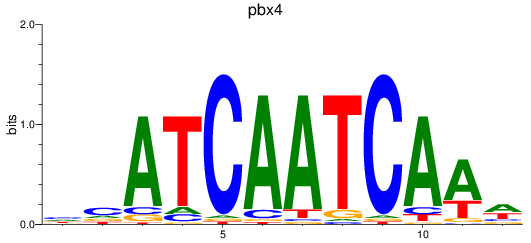
Results for pdx1_pbx4
Z-value: 0.88
Transcription factors associated with pdx1_pbx4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pdx1
|
ENSDARG00000002779 | pancreatic and duodenal homeobox 1 |
|
pdx1
|
ENSDARG00000109275 | pancreatic and duodenal homeobox 1 |
|
pbx4
|
ENSDARG00000052150 | pre-B-cell leukemia transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pbx4 | dr11_v1_chr3_+_53052769_53052769 | 0.25 | 3.1e-01 | Click! |
| pdx1 | dr11_v1_chr24_+_21684189_21684189 | -0.20 | 4.1e-01 | Click! |
Activity profile of pdx1_pbx4 motif
Sorted Z-values of pdx1_pbx4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_36243774 | 5.31 |
ENSDART00000023323
|
zgc:172270
|
zgc:172270 |
| chr12_-_11457625 | 3.70 |
ENSDART00000012318
|
htra1b
|
HtrA serine peptidase 1b |
| chr18_+_13792490 | 3.65 |
ENSDART00000136754
|
cdh13
|
cadherin 13, H-cadherin (heart) |
| chr24_+_38301080 | 3.44 |
ENSDART00000105672
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr20_+_1088658 | 3.05 |
ENSDART00000162991
|
BX537249.1
|
|
| chr14_+_35237613 | 2.96 |
ENSDART00000163465
|
ebf3a
|
early B cell factor 3a |
| chr24_-_6898302 | 2.56 |
ENSDART00000158646
|
dpp6a
|
dipeptidyl-peptidase 6a |
| chr4_-_16333944 | 2.01 |
ENSDART00000079523
|
epyc
|
epiphycan |
| chr1_-_26023678 | 1.95 |
ENSDART00000054202
|
si:ch211-145b13.5
|
si:ch211-145b13.5 |
| chr12_+_39685485 | 1.92 |
ENSDART00000163403
|
LO017650.1
|
|
| chr1_+_17676745 | 1.91 |
ENSDART00000030665
|
slc25a4
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 |
| chr24_-_6897884 | 1.87 |
ENSDART00000080766
|
dpp6a
|
dipeptidyl-peptidase 6a |
| chr8_+_6954984 | 1.67 |
ENSDART00000145610
|
si:ch211-255g12.6
|
si:ch211-255g12.6 |
| chr7_-_35066457 | 1.64 |
ENSDART00000058067
|
zgc:112160
|
zgc:112160 |
| chr4_+_21129752 | 1.63 |
ENSDART00000169764
|
syt1a
|
synaptotagmin Ia |
| chr15_-_27710513 | 1.57 |
ENSDART00000005641
ENSDART00000134373 |
lhx1a
|
LIM homeobox 1a |
| chr23_-_6641223 | 1.55 |
ENSDART00000023793
|
mipb
|
major intrinsic protein of lens fiber b |
| chr15_+_4632782 | 1.54 |
ENSDART00000156012
|
si:dkey-35i13.1
|
si:dkey-35i13.1 |
| chr18_+_783936 | 1.49 |
ENSDART00000193357
|
rpp25b
|
ribonuclease P and MRP subunit p25, b |
| chr19_+_37925616 | 1.48 |
ENSDART00000148348
|
nxph1
|
neurexophilin 1 |
| chr20_-_8443425 | 1.41 |
ENSDART00000083908
|
dab1a
|
Dab, reelin signal transducer, homolog 1a (Drosophila) |
| chr2_+_2223837 | 1.35 |
ENSDART00000101038
ENSDART00000129354 |
tmie
|
transmembrane inner ear |
| chr21_-_43079161 | 1.33 |
ENSDART00000144151
|
jakmip2
|
janus kinase and microtubule interacting protein 2 |
| chr5_-_29643930 | 1.32 |
ENSDART00000161250
|
grin1b
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1b |
| chr22_+_3223489 | 1.29 |
ENSDART00000082011
|
lim2.2
|
lens intrinsic membrane protein 2.2 |
| chr22_+_35068046 | 1.23 |
ENSDART00000161660
ENSDART00000169573 |
si:ch73-173h19.3
|
si:ch73-173h19.3 |
| chr16_-_16152199 | 1.18 |
ENSDART00000012718
|
fabp11b
|
fatty acid binding protein 11b |
| chr9_+_42066030 | 1.16 |
ENSDART00000185311
ENSDART00000015267 |
pcbp3
|
poly(rC) binding protein 3 |
| chr10_-_26744131 | 1.13 |
ENSDART00000020096
ENSDART00000162710 ENSDART00000179853 |
fgf13b
|
fibroblast growth factor 13b |
| chr12_-_26406323 | 1.12 |
ENSDART00000131896
|
myoz1b
|
myozenin 1b |
| chr9_-_23217196 | 1.09 |
ENSDART00000083567
|
kif5c
|
kinesin family member 5C |
| chr1_-_43963441 | 1.06 |
ENSDART00000137646
|
odam
|
odontogenic, ameloblast associated |
| chr19_-_5345930 | 1.05 |
ENSDART00000066620
ENSDART00000151398 |
krtt1c19e
|
keratin type 1 c19e |
| chr7_-_8374950 | 1.03 |
ENSDART00000057101
|
aep1
|
aerolysin-like protein |
| chr14_-_2199573 | 1.03 |
ENSDART00000124485
|
pcdh2ab8
|
protocadherin 2 alpha b 8 |
| chr5_-_50992690 | 1.01 |
ENSDART00000149553
ENSDART00000097460 ENSDART00000192021 |
hmgcra
|
3-hydroxy-3-methylglutaryl-CoA reductase a |
| chr12_+_16440708 | 1.00 |
ENSDART00000113810
|
ankrd1b
|
ankyrin repeat domain 1b (cardiac muscle) |
| chr19_-_26869103 | 0.99 |
ENSDART00000089699
|
prrt1
|
proline-rich transmembrane protein 1 |
| chr19_-_10395683 | 0.98 |
ENSDART00000109488
|
zgc:194578
|
zgc:194578 |
| chr25_+_20215964 | 0.94 |
ENSDART00000139235
|
tnnt2d
|
troponin T2d, cardiac |
| chr20_-_29420713 | 0.92 |
ENSDART00000147464
|
ryr3
|
ryanodine receptor 3 |
| chr4_-_4387012 | 0.91 |
ENSDART00000191836
|
CU468826.3
|
Danio rerio U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit-related protein 1-like (LOC100331497), mRNA. |
| chr12_+_27213733 | 0.90 |
ENSDART00000133048
|
nbr1a
|
neighbor of brca1 gene 1a |
| chr12_+_38770654 | 0.90 |
ENSDART00000155367
|
kif19
|
kinesin family member 19 |
| chr25_+_15354095 | 0.89 |
ENSDART00000090397
|
kiaa1549la
|
KIAA1549-like a |
| chr2_+_42724404 | 0.89 |
ENSDART00000075392
|
basp1
|
brain abundant, membrane attached signal protein 1 |
| chr7_-_52842007 | 0.89 |
ENSDART00000182710
|
map1aa
|
microtubule-associated protein 1Aa |
| chr8_+_22931427 | 0.89 |
ENSDART00000063096
|
sypa
|
synaptophysin a |
| chr12_+_9703172 | 0.88 |
ENSDART00000091489
|
ppp1r9bb
|
protein phosphatase 1, regulatory subunit 9Bb |
| chr18_-_38088099 | 0.86 |
ENSDART00000146120
|
luzp2
|
leucine zipper protein 2 |
| chr8_+_44703864 | 0.85 |
ENSDART00000016225
|
star
|
steroidogenic acute regulatory protein |
| chr2_-_722156 | 0.84 |
ENSDART00000045770
ENSDART00000169498 |
foxq1a
|
forkhead box Q1a |
| chr7_+_46252993 | 0.83 |
ENSDART00000167149
|
znf536
|
zinc finger protein 536 |
| chr4_-_27301356 | 0.83 |
ENSDART00000100444
|
fam19a5a
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5a |
| chr6_-_46875310 | 0.82 |
ENSDART00000154442
|
igfn1.3
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 3 |
| chr25_+_20216159 | 0.82 |
ENSDART00000048642
|
tnnt2d
|
troponin T2d, cardiac |
| chr24_-_40009446 | 0.82 |
ENSDART00000087422
|
aoc1
|
amine oxidase, copper containing 1 |
| chr7_-_8417315 | 0.81 |
ENSDART00000173046
|
jac1
|
jacalin 1 |
| chr5_+_44654535 | 0.81 |
ENSDART00000182190
ENSDART00000181872 |
dapk1
|
death-associated protein kinase 1 |
| chr16_+_10776688 | 0.80 |
ENSDART00000161969
ENSDART00000172657 |
atp1a3b
|
ATPase Na+/K+ transporting subunit alpha 3b |
| chr8_-_34052019 | 0.80 |
ENSDART00000040126
ENSDART00000159208 ENSDART00000048994 ENSDART00000098822 |
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr5_-_51619742 | 0.79 |
ENSDART00000188537
|
otpb
|
orthopedia homeobox b |
| chr4_-_5291256 | 0.78 |
ENSDART00000150864
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr5_-_23280098 | 0.78 |
ENSDART00000126540
ENSDART00000051533 |
plp1b
|
proteolipid protein 1b |
| chr4_-_4780667 | 0.77 |
ENSDART00000133973
|
si:ch211-258f14.2
|
si:ch211-258f14.2 |
| chr3_-_32818607 | 0.76 |
ENSDART00000075465
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr18_-_42172101 | 0.75 |
ENSDART00000124211
|
cntn5
|
contactin 5 |
| chr25_-_29363934 | 0.74 |
ENSDART00000166889
|
nptna
|
neuroplastin a |
| chr11_-_44030962 | 0.73 |
ENSDART00000171910
|
FP016005.1
|
|
| chr10_-_39011514 | 0.70 |
ENSDART00000075123
|
pcp4a
|
Purkinje cell protein 4a |
| chr1_-_19845378 | 0.68 |
ENSDART00000139314
ENSDART00000132958 ENSDART00000147502 |
grhprb
|
glyoxylate reductase/hydroxypyruvate reductase b |
| chr3_-_52644737 | 0.68 |
ENSDART00000126180
|
si:dkey-210j14.3
|
si:dkey-210j14.3 |
| chr9_+_2002701 | 0.68 |
ENSDART00000082329
|
evx2
|
even-skipped homeobox 2 |
| chr13_+_38302665 | 0.67 |
ENSDART00000145777
|
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr7_+_73593814 | 0.66 |
ENSDART00000110544
|
znf219
|
zinc finger protein 219 |
| chr10_+_439692 | 0.65 |
ENSDART00000147740
|
zdhhc8a
|
zinc finger, DHHC-type containing 8a |
| chr21_+_40092301 | 0.64 |
ENSDART00000145150
|
serpinf2a
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2a |
| chr25_-_4482449 | 0.64 |
ENSDART00000056278
ENSDART00000149425 |
slc25a22a
|
solute carrier family 25 member 22a |
| chr12_-_20362041 | 0.64 |
ENSDART00000184145
ENSDART00000105952 |
aqp8a.2
|
aquaporin 8a, tandem duplicate 2 |
| chr12_-_26064480 | 0.63 |
ENSDART00000158215
ENSDART00000171206 ENSDART00000171212 ENSDART00000182956 ENSDART00000186779 |
ldb3b
|
LIM domain binding 3b |
| chr1_-_46981134 | 0.63 |
ENSDART00000130607
|
pknox1.2
|
pbx/knotted 1 homeobox 1.2 |
| chr3_-_20040636 | 0.62 |
ENSDART00000104118
|
atxn7l3
|
ataxin 7-like 3 |
| chr3_-_16068236 | 0.62 |
ENSDART00000157315
|
si:dkey-81l17.6
|
si:dkey-81l17.6 |
| chr12_-_28881638 | 0.60 |
ENSDART00000148459
ENSDART00000039667 ENSDART00000148668 ENSDART00000136593 ENSDART00000139923 ENSDART00000148912 |
cbx1b
|
chromobox homolog 1b (HP1 beta homolog Drosophila) |
| chr4_-_42408339 | 0.60 |
ENSDART00000172612
|
si:ch211-59d8.3
|
si:ch211-59d8.3 |
| chr17_-_12385308 | 0.60 |
ENSDART00000080927
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr23_-_11870962 | 0.59 |
ENSDART00000143481
|
si:dkey-178k16.1
|
si:dkey-178k16.1 |
| chr21_+_30194904 | 0.59 |
ENSDART00000137023
ENSDART00000078403 |
BRD8
|
si:ch211-59d17.3 |
| chr25_-_30047477 | 0.59 |
ENSDART00000164859
|
CR759810.1
|
|
| chr25_+_13406069 | 0.59 |
ENSDART00000010495
|
znrf1
|
zinc and ring finger 1 |
| chr23_+_5490854 | 0.58 |
ENSDART00000175403
|
tulp1a
|
tubby like protein 1a |
| chr25_-_7925269 | 0.58 |
ENSDART00000014274
|
glcea
|
glucuronic acid epimerase a |
| chr19_-_10243148 | 0.58 |
ENSDART00000148073
|
shisa7b
|
shisa family member 7 |
| chr20_-_29418620 | 0.57 |
ENSDART00000172634
|
ryr3
|
ryanodine receptor 3 |
| chr3_+_32862730 | 0.57 |
ENSDART00000144939
ENSDART00000125126 ENSDART00000144531 ENSDART00000141717 ENSDART00000137599 |
zgc:162613
|
zgc:162613 |
| chr9_-_33725972 | 0.57 |
ENSDART00000028225
|
mao
|
monoamine oxidase |
| chr1_-_19215336 | 0.56 |
ENSDART00000162949
ENSDART00000170680 |
ptprdb
|
protein tyrosine phosphatase, receptor type, D, b |
| chr6_+_40661703 | 0.55 |
ENSDART00000142492
|
eno1b
|
enolase 1b, (alpha) |
| chr23_+_4689626 | 0.55 |
ENSDART00000131532
|
gp9
|
glycoprotein IX (platelet) |
| chr1_-_681116 | 0.55 |
ENSDART00000165894
|
adamts1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr7_-_13882988 | 0.55 |
ENSDART00000169828
|
rlbp1a
|
retinaldehyde binding protein 1a |
| chr13_+_22479988 | 0.55 |
ENSDART00000188182
ENSDART00000192972 ENSDART00000178372 |
ldb3a
|
LIM domain binding 3a |
| chr4_+_26496489 | 0.54 |
ENSDART00000160652
|
iqsec3a
|
IQ motif and Sec7 domain 3a |
| chr19_+_4443285 | 0.54 |
ENSDART00000162683
|
trappc9
|
trafficking protein particle complex 9 |
| chr12_+_48634927 | 0.54 |
ENSDART00000168441
|
zgc:165653
|
zgc:165653 |
| chr2_+_7818368 | 0.54 |
ENSDART00000007068
|
kcnmb2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr18_+_41495841 | 0.54 |
ENSDART00000098671
|
si:ch211-203b8.6
|
si:ch211-203b8.6 |
| chr24_-_24881996 | 0.54 |
ENSDART00000145712
|
crhb
|
corticotropin releasing hormone b |
| chr23_+_40460333 | 0.54 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr16_-_28658341 | 0.53 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr4_+_40427249 | 0.53 |
ENSDART00000151927
|
si:ch211-218h8.3
|
si:ch211-218h8.3 |
| chr24_+_32472155 | 0.53 |
ENSDART00000098859
|
neurod6a
|
neuronal differentiation 6a |
| chr25_-_7925019 | 0.53 |
ENSDART00000183309
|
glcea
|
glucuronic acid epimerase a |
| chr8_+_21376290 | 0.52 |
ENSDART00000136765
|
ela2
|
elastase 2 |
| chr21_-_27338639 | 0.52 |
ENSDART00000130632
|
hif1al2
|
hypoxia-inducible factor 1, alpha subunit, like 2 |
| chr17_+_23937262 | 0.52 |
ENSDART00000113276
|
si:ch211-189k9.2
|
si:ch211-189k9.2 |
| chr4_+_13810811 | 0.50 |
ENSDART00000067168
|
pdzrn4
|
PDZ domain containing ring finger 4 |
| chr18_-_36135799 | 0.50 |
ENSDART00000059344
|
b3gat1a
|
beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P) a |
| chr16_+_46453176 | 0.49 |
ENSDART00000132793
|
rpz3
|
rapunzel 3 |
| chr23_+_28322986 | 0.49 |
ENSDART00000134710
|
birc5b
|
baculoviral IAP repeat containing 5b |
| chr17_+_9308425 | 0.49 |
ENSDART00000188283
ENSDART00000183311 |
NPAS3
|
neuronal PAS domain protein 3 |
| chr22_-_33679277 | 0.49 |
ENSDART00000169948
|
FO904977.1
|
|
| chr8_+_21406769 | 0.48 |
ENSDART00000135766
|
si:dkey-163f12.6
|
si:dkey-163f12.6 |
| chr6_+_4872883 | 0.48 |
ENSDART00000186730
ENSDART00000092290 ENSDART00000151674 |
pcdh9
|
protocadherin 9 |
| chr1_-_23557877 | 0.48 |
ENSDART00000145942
|
fam184b
|
family with sequence similarity 184, member B |
| chr4_+_7391110 | 0.47 |
ENSDART00000160708
ENSDART00000187823 |
tnni4a
|
troponin I4a |
| chr13_-_33000649 | 0.47 |
ENSDART00000133677
|
rbm25a
|
RNA binding motif protein 25a |
| chr4_-_60314809 | 0.46 |
ENSDART00000164606
|
si:dkey-248e17.4
|
si:dkey-248e17.4 |
| chr13_+_22480496 | 0.46 |
ENSDART00000136863
ENSDART00000131870 ENSDART00000078720 ENSDART00000078740 ENSDART00000139218 |
ldb3a
|
LIM domain binding 3a |
| chr2_-_10188598 | 0.46 |
ENSDART00000189122
|
dmbx1a
|
diencephalon/mesencephalon homeobox 1a |
| chr6_+_45347219 | 0.46 |
ENSDART00000188240
|
LO017951.1
|
|
| chr18_-_20466061 | 0.46 |
ENSDART00000060311
|
paqr5a
|
progestin and adipoQ receptor family member Va |
| chr1_-_26294995 | 0.46 |
ENSDART00000168594
|
cxxc4
|
CXXC finger 4 |
| chr8_+_23165749 | 0.46 |
ENSDART00000063057
|
dnajc5aa
|
DnaJ (Hsp40) homolog, subfamily C, member 5aa |
| chr13_+_50375800 | 0.46 |
ENSDART00000099537
|
cox5b2
|
cytochrome c oxidase subunit Vb 2 |
| chr17_-_16133249 | 0.45 |
ENSDART00000030919
|
pnoca
|
prepronociceptin a |
| chr14_+_36231126 | 0.45 |
ENSDART00000141766
|
elovl6
|
ELOVL fatty acid elongase 6 |
| chr2_+_45068366 | 0.44 |
ENSDART00000142175
|
si:dkey-76d14.2
|
si:dkey-76d14.2 |
| chr7_+_29952719 | 0.44 |
ENSDART00000173737
|
tpma
|
alpha-tropomyosin |
| chr9_-_22834860 | 0.43 |
ENSDART00000146486
|
neb
|
nebulin |
| chr14_+_3495542 | 0.43 |
ENSDART00000168934
|
gstp2
|
glutathione S-transferase pi 2 |
| chr21_+_5518619 | 0.43 |
ENSDART00000170587
|
ly6m5
|
lymphocyte antigen 6 family member M5 |
| chr6_+_45346808 | 0.43 |
ENSDART00000186327
|
LO017951.1
|
|
| chr8_+_38415374 | 0.42 |
ENSDART00000085395
|
nkx6.3
|
NK6 homeobox 3 |
| chr8_+_44420108 | 0.42 |
ENSDART00000075381
|
CU571323.1
|
|
| chr13_+_22249636 | 0.41 |
ENSDART00000108472
ENSDART00000173123 |
synpo2la
|
synaptopodin 2-like a |
| chr17_+_19626479 | 0.41 |
ENSDART00000044993
ENSDART00000131863 |
rgs7a
|
regulator of G protein signaling 7a |
| chr14_-_4044545 | 0.41 |
ENSDART00000169527
|
snx25
|
sorting nexin 25 |
| chr20_+_4793790 | 0.41 |
ENSDART00000153486
|
lgals8a
|
galectin 8a |
| chr2_+_26647472 | 0.41 |
ENSDART00000145415
ENSDART00000157409 |
ttpa
|
tocopherol (alpha) transfer protein |
| chr5_+_9382301 | 0.41 |
ENSDART00000124017
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr3_-_59981476 | 0.40 |
ENSDART00000035878
ENSDART00000124038 |
cdr2l
|
cerebellar degeneration-related protein 2-like |
| chr25_-_13842618 | 0.40 |
ENSDART00000160258
|
mapk8ip1a
|
mitogen-activated protein kinase 8 interacting protein 1a |
| chr6_-_47246948 | 0.40 |
ENSDART00000162435
|
grm4
|
glutamate receptor, metabotropic 4 |
| chr12_-_31009315 | 0.40 |
ENSDART00000133854
|
tcf7l2
|
transcription factor 7 like 2 |
| chr11_-_17713987 | 0.40 |
ENSDART00000090401
|
fam19a4b
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A4b |
| chr1_-_40227166 | 0.40 |
ENSDART00000146680
|
si:ch211-113e8.3
|
si:ch211-113e8.3 |
| chr2_+_25657958 | 0.39 |
ENSDART00000161407
|
tnika
|
TRAF2 and NCK interacting kinase a |
| chr2_-_18830722 | 0.39 |
ENSDART00000165330
ENSDART00000165698 |
pbx1a
|
pre-B-cell leukemia homeobox 1a |
| chr23_+_216012 | 0.39 |
ENSDART00000181115
ENSDART00000004678 ENSDART00000190439 ENSDART00000189322 |
PDZD4
|
si:ch73-162j3.4 |
| chr17_-_6399920 | 0.39 |
ENSDART00000022010
|
hivep2b
|
human immunodeficiency virus type I enhancer binding protein 2b |
| chr7_-_8408014 | 0.39 |
ENSDART00000112492
|
zgc:194686
|
zgc:194686 |
| chr14_+_34547554 | 0.39 |
ENSDART00000074819
|
gabrp
|
gamma-aminobutyric acid (GABA) A receptor, pi |
| chr7_+_26629084 | 0.39 |
ENSDART00000101044
ENSDART00000173765 |
hsbp1a
|
heat shock factor binding protein 1a |
| chr15_+_30917282 | 0.38 |
ENSDART00000129474
|
omgb
|
oligodendrocyte myelin glycoprotein b |
| chr1_-_46984142 | 0.37 |
ENSDART00000125032
|
pknox1.2
|
pbx/knotted 1 homeobox 1.2 |
| chr11_+_24002503 | 0.37 |
ENSDART00000164702
|
chia.2
|
chitinase, acidic.2 |
| chr17_+_39242437 | 0.37 |
ENSDART00000156138
ENSDART00000128863 |
zgc:174356
|
zgc:174356 |
| chr6_-_53143667 | 0.37 |
ENSDART00000079694
|
gnai2b
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2b |
| chr2_-_716426 | 0.36 |
ENSDART00000028159
|
foxf2a
|
forkhead box F2a |
| chr13_+_23282549 | 0.36 |
ENSDART00000101134
|
khdrbs2
|
KH domain containing, RNA binding, signal transduction associated 2 |
| chr17_-_49800869 | 0.36 |
ENSDART00000156264
|
col12a1a
|
collagen, type XII, alpha 1a |
| chr17_-_30666037 | 0.36 |
ENSDART00000156509
|
alkal2b
|
ALK and LTK ligand 2b |
| chr10_+_21511495 | 0.36 |
ENSDART00000178395
|
BX957322.3
|
|
| chr24_-_22533959 | 0.36 |
ENSDART00000148197
|
ctnnd2a
|
catenin (cadherin-associated protein), delta 2a |
| chr21_+_25181003 | 0.35 |
ENSDART00000169700
|
si:dkey-183i3.9
|
si:dkey-183i3.9 |
| chr17_-_26507289 | 0.35 |
ENSDART00000155616
|
ccser2a
|
coiled-coil serine-rich protein 2a |
| chr10_+_26571174 | 0.35 |
ENSDART00000148617
ENSDART00000112956 |
slc9a6b
|
solute carrier family 9, subfamily A (NHE6, cation proton antiporter 6), member 6b |
| chr6_-_49510553 | 0.35 |
ENSDART00000166238
|
rplp2
|
ribosomal protein, large P2 |
| chr14_-_45702721 | 0.35 |
ENSDART00000165727
|
si:dkey-226n24.1
|
si:dkey-226n24.1 |
| chr1_+_9144958 | 0.35 |
ENSDART00000046507
|
rasd3
|
RASD family member 3 |
| chr1_+_47165842 | 0.35 |
ENSDART00000053152
ENSDART00000167051 |
cbr1
|
carbonyl reductase 1 |
| chr4_-_75678082 | 0.35 |
ENSDART00000180201
|
si:dkey-165o8.1
|
si:dkey-165o8.1 |
| chr2_+_53357953 | 0.34 |
ENSDART00000187577
|
celf5b
|
cugbp, Elav-like family member 5b |
| chr6_-_15641686 | 0.34 |
ENSDART00000135583
|
mlpha
|
melanophilin a |
| chr4_+_13640903 | 0.34 |
ENSDART00000155349
|
pnpla8
|
patatin-like phospholipase domain containing 8 |
| chr10_-_17745345 | 0.34 |
ENSDART00000132690
ENSDART00000135376 |
si:dkey-200l5.4
|
si:dkey-200l5.4 |
| chr19_+_25478203 | 0.34 |
ENSDART00000132934
|
col28a1a
|
collagen, type XXVIII, alpha 1a |
| chr9_-_44939104 | 0.34 |
ENSDART00000192903
|
vil1
|
villin 1 |
| chr14_+_51056605 | 0.34 |
ENSDART00000159639
|
CABZ01078593.1
|
|
| chr8_-_39903803 | 0.33 |
ENSDART00000012391
|
cabp1a
|
calcium binding protein 1a |
| chr22_-_16180849 | 0.33 |
ENSDART00000090390
|
vcam1b
|
vascular cell adhesion molecule 1b |
| chr7_+_35229805 | 0.33 |
ENSDART00000173911
|
tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr23_-_21515182 | 0.33 |
ENSDART00000142000
|
rnf207b
|
ring finger protein 207b |
| chr13_+_24750078 | 0.33 |
ENSDART00000021053
|
col17a1b
|
collagen, type XVII, alpha 1b |
| chr24_+_26402110 | 0.33 |
ENSDART00000133684
|
si:ch211-230g15.5
|
si:ch211-230g15.5 |
| chr2_+_5948534 | 0.33 |
ENSDART00000124324
ENSDART00000176461 |
slc1a7a
|
solute carrier family 1 (glutamate transporter), member 7a |
| chr2_+_37480669 | 0.32 |
ENSDART00000029801
|
sppl2
|
signal peptide peptidase-like 2 |
| chr11_-_6048490 | 0.32 |
ENSDART00000066164
|
plvapb
|
plasmalemma vesicle associated protein b |
| chr5_+_9377005 | 0.32 |
ENSDART00000124924
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of pdx1_pbx4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 5.3 | GO:0070836 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.5 | 1.6 | GO:0090184 | regulation of morphogenesis of a branching structure(GO:0060688) positive regulation of mesonephros development(GO:0061213) regulation of mesonephros development(GO:0061217) regulation of kidney development(GO:0090183) positive regulation of kidney development(GO:0090184) regulation of branching involved in ureteric bud morphogenesis(GO:0090189) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) interneuron axon guidance(GO:0097376) spinal cord interneuron axon guidance(GO:0097377) dorsal spinal cord interneuron axon guidance(GO:0097378) |
| 0.5 | 1.9 | GO:0015867 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.5 | 3.6 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.2 | 1.4 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.2 | 0.5 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.2 | 1.6 | GO:0006833 | water transport(GO:0006833) |
| 0.2 | 1.5 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.2 | 0.5 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.2 | 0.8 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.2 | 1.1 | GO:0098935 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.1 | 4.6 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.1 | 0.8 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.1 | 0.7 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.1 | 0.8 | GO:0021767 | mammillary body development(GO:0021767) |
| 0.1 | 1.4 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.4 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.1 | 1.1 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.1 | 0.3 | GO:1990416 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.1 | 0.5 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 0.5 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.1 | 0.3 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.1 | 0.3 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) regulation of lamellipodium morphogenesis(GO:2000392) |
| 0.1 | 1.6 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 1.3 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 0.4 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.1 | 1.3 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 0.6 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 0.3 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 0.6 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 0.6 | GO:0043490 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.6 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.1 | 0.7 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 0.4 | GO:0070376 | ERK5 cascade(GO:0070375) regulation of ERK5 cascade(GO:0070376) positive regulation of ERK5 cascade(GO:0070378) |
| 0.1 | 0.3 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.1 | 0.2 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.1 | 1.0 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.1 | 0.3 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 0.2 | GO:0060300 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.0 | 0.5 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.2 | GO:1902514 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.0 | 1.6 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.0 | 0.2 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.2 | GO:0018199 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.2 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.0 | 0.3 | GO:0071233 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.0 | 0.3 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.8 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.2 | GO:0060547 | negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.0 | 0.4 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.4 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 1.8 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.0 | 0.6 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.0 | 0.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.0 | 0.2 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) |
| 0.0 | 1.6 | GO:0031102 | neuron projection regeneration(GO:0031102) |
| 0.0 | 0.9 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.2 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.4 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.0 | 0.1 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 1.0 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.3 | GO:0016119 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.0 | 0.3 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 0.1 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.0 | 0.3 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.2 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.9 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.2 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.1 | GO:0048903 | anterior lateral line neuromast hair cell differentiation(GO:0048903) |
| 0.0 | 0.2 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.1 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 1.0 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.4 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.6 | GO:0051492 | regulation of stress fiber assembly(GO:0051492) |
| 0.0 | 1.2 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 0.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.4 | GO:0006775 | fat-soluble vitamin metabolic process(GO:0006775) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.3 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.0 | 0.4 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.5 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.0 | 0.1 | GO:0010991 | SMAD protein complex assembly(GO:0007183) regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:0090179 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.6 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.6 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 0.3 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.4 | GO:0036294 | cellular response to decreased oxygen levels(GO:0036294) cellular response to hypoxia(GO:0071456) |
| 0.0 | 0.1 | GO:0048899 | anterior lateral line development(GO:0048899) |
| 0.0 | 0.1 | GO:0003245 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.0 | 0.1 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.1 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.6 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.3 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.1 | GO:0070291 | N-acylethanolamine metabolic process(GO:0070291) |
| 0.0 | 0.4 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.2 | GO:0034394 | protein localization to cell surface(GO:0034394) energy homeostasis(GO:0097009) |
| 0.0 | 0.3 | GO:1900153 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 0.7 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.2 | GO:0016198 | axon choice point recognition(GO:0016198) axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.2 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.1 | GO:0002507 | tolerance induction(GO:0002507) tolerance induction to self antigen(GO:0002513) regulation of platelet activation(GO:0010543) regulation of platelet aggregation(GO:0090330) |
| 0.0 | 0.5 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.1 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.4 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.2 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.3 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.2 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.0 | 0.7 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 1.1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.2 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.3 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.2 | GO:0099587 | inorganic cation import into cell(GO:0098659) inorganic ion import into cell(GO:0099587) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.2 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.1 | GO:0002504 | antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.0 | 0.3 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.2 | 1.5 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.2 | 1.5 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 5.3 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 0.7 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 0.6 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 0.4 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.1 | 1.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.3 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 0.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.3 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 5.0 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 2.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 2.8 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.3 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 1.0 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.8 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.3 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 1.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.6 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.2 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 0.8 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 1.7 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.4 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.3 | GO:0032589 | neuron projection membrane(GO:0032589) dendrite membrane(GO:0032590) |
| 0.0 | 0.8 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.4 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.1 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 0.7 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.7 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.0 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 1.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.5 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 1.1 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.1 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 1.4 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 1.2 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.8 | GO:0048786 | presynaptic active zone(GO:0048786) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.4 | 1.3 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.3 | 1.0 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.3 | 1.5 | GO:0048763 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.3 | 1.1 | GO:0047464 | heparosan-N-sulfate-glucuronate 5-epimerase activity(GO:0047464) |
| 0.3 | 1.4 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.2 | 1.0 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.2 | 4.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.2 | 2.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.2 | 0.7 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.2 | 0.5 | GO:0015562 | protein-transmembrane transporting ATPase activity(GO:0015462) efflux transmembrane transporter activity(GO:0015562) |
| 0.2 | 1.8 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.1 | 3.7 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 0.5 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.1 | 1.1 | GO:0031433 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.1 | 0.3 | GO:0060175 | brain-derived neurotrophic factor binding(GO:0048403) brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.1 | 0.8 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 1.6 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 0.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 1.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 0.7 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.1 | 0.9 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.4 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.1 | 0.6 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 1.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 1.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.5 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.2 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.2 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.0 | 0.3 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.3 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.0 | 0.2 | GO:0031781 | type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.3 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.5 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.4 | GO:0015385 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 1.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 3.3 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.3 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.2 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.0 | 0.5 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.4 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.2 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.0 | 1.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.3 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 0.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.2 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.0 | 0.2 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.2 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.4 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.6 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.4 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.3 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.1 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 3.7 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 0.4 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 1.1 | GO:1902936 | phosphatidylinositol bisphosphate binding(GO:1902936) |
| 0.0 | 0.4 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 1.0 | GO:0050661 | NADP binding(GO:0050661) |
| 0.0 | 0.2 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.2 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.5 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.3 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.9 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.2 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.7 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0047192 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.2 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.2 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 1.2 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.6 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.0 | GO:0008119 | thiopurine S-methyltransferase activity(GO:0008119) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.8 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.2 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 0.8 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 0.6 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 0.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 1.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 0.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.6 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 1.0 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.0 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.1 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |