Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
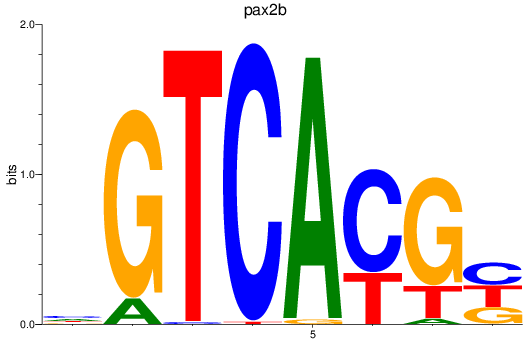
Results for pax2b
Z-value: 0.48
Transcription factors associated with pax2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pax2b
|
ENSDARG00000032578 | paired box 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pax2b | dr11_v1_chr12_-_45875946_45875946 | -0.11 | 6.4e-01 | Click! |
Activity profile of pax2b motif
Sorted Z-values of pax2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_24758967 | 0.65 |
ENSDART00000005616
ENSDART00000133481 |
rnpep
|
arginyl aminopeptidase (aminopeptidase B) |
| chr3_+_33300522 | 0.61 |
ENSDART00000114023
|
hspb9
|
heat shock protein, alpha-crystallin-related, 9 |
| chr20_-_25626693 | 0.55 |
ENSDART00000132247
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr20_-_25626198 | 0.53 |
ENSDART00000126716
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr22_-_25612680 | 0.52 |
ENSDART00000114167
|
si:ch211-12h2.8
|
si:ch211-12h2.8 |
| chr20_-_25626428 | 0.51 |
ENSDART00000136475
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr19_+_15441022 | 0.51 |
ENSDART00000098970
ENSDART00000140276 |
lin28a
|
lin-28 homolog A (C. elegans) |
| chr22_-_11136625 | 0.48 |
ENSDART00000016873
ENSDART00000125561 |
atp6ap2
|
ATPase H+ transporting accessory protein 2 |
| chr17_-_114121 | 0.44 |
ENSDART00000172408
ENSDART00000157784 |
arhgap11a
|
Rho GTPase activating protein 11A |
| chr23_-_21471022 | 0.44 |
ENSDART00000104206
|
her4.2
|
hairy-related 4, tandem duplicate 2 |
| chr3_-_21062706 | 0.43 |
ENSDART00000155605
ENSDART00000153686 ENSDART00000157168 ENSDART00000156614 ENSDART00000155743 ENSDART00000156275 |
fam57ba
|
family with sequence similarity 57, member Ba |
| chr3_-_37785873 | 0.42 |
ENSDART00000011691
|
baxa
|
BCL2 associated X, apoptosis regulator a |
| chr2_+_30481125 | 0.41 |
ENSDART00000125933
|
fam173b
|
family with sequence similarity 173, member B |
| chr23_+_31913292 | 0.41 |
ENSDART00000136910
|
armc1l
|
armadillo repeat containing 1, like |
| chr14_+_97017 | 0.41 |
ENSDART00000159300
ENSDART00000169523 |
mcm7
|
minichromosome maintenance complex component 7 |
| chr19_-_47570672 | 0.41 |
ENSDART00000112155
|
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr11_+_13176568 | 0.40 |
ENSDART00000125371
ENSDART00000123257 |
mknk1
|
MAP kinase interacting serine/threonine kinase 1 |
| chr6_-_55297274 | 0.40 |
ENSDART00000184283
|
ube2c
|
ubiquitin-conjugating enzyme E2C |
| chr13_+_25449681 | 0.40 |
ENSDART00000101328
|
atoh7
|
atonal bHLH transcription factor 7 |
| chr3_+_34670076 | 0.39 |
ENSDART00000133457
|
dlx4a
|
distal-less homeobox 4a |
| chr7_-_28696556 | 0.39 |
ENSDART00000148822
|
adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr1_-_55166511 | 0.38 |
ENSDART00000150430
ENSDART00000035725 |
pane1
|
proliferation associated nuclear element |
| chr3_-_26017592 | 0.38 |
ENSDART00000030890
|
hmox1a
|
heme oxygenase 1a |
| chr23_+_44644911 | 0.38 |
ENSDART00000140799
|
zgc:85858
|
zgc:85858 |
| chr3_-_26017831 | 0.38 |
ENSDART00000179982
|
hmox1a
|
heme oxygenase 1a |
| chr8_-_41228530 | 0.38 |
ENSDART00000165949
ENSDART00000173055 |
fahd2a
|
fumarylacetoacetate hydrolase domain containing 2A |
| chr3_-_32362872 | 0.36 |
ENSDART00000035545
ENSDART00000012630 |
prmt1
|
protein arginine methyltransferase 1 |
| chr13_-_35892243 | 0.36 |
ENSDART00000002750
ENSDART00000122810 ENSDART00000162399 |
tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr13_-_45022301 | 0.35 |
ENSDART00000183589
ENSDART00000125633 ENSDART00000074787 |
khdrbs1a
|
KH domain containing, RNA binding, signal transduction associated 1a |
| chr3_+_39566999 | 0.35 |
ENSDART00000146867
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr10_+_23060391 | 0.35 |
ENSDART00000079711
|
slc25a1a
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1a |
| chr5_-_67365006 | 0.35 |
ENSDART00000136116
|
unga
|
uracil DNA glycosylase a |
| chr3_+_23768898 | 0.35 |
ENSDART00000110682
|
hoxb1a
|
homeobox B1a |
| chr6_-_21534301 | 0.35 |
ENSDART00000126186
|
psmd12
|
proteasome 26S subunit, non-ATPase 12 |
| chr21_-_21465111 | 0.35 |
ENSDART00000141487
|
nectin3b
|
nectin cell adhesion molecule 3b |
| chr4_-_16836006 | 0.35 |
ENSDART00000010777
|
ldhba
|
lactate dehydrogenase Ba |
| chr10_+_36662640 | 0.34 |
ENSDART00000063359
|
ucp2
|
uncoupling protein 2 |
| chr11_-_22372072 | 0.34 |
ENSDART00000065996
|
tmem183a
|
transmembrane protein 183A |
| chr25_+_19008497 | 0.34 |
ENSDART00000104420
|
samm50
|
SAMM50 sorting and assembly machinery component |
| chr23_-_1056808 | 0.34 |
ENSDART00000081961
|
zgc:113423
|
zgc:113423 |
| chr2_+_30480907 | 0.33 |
ENSDART00000041378
ENSDART00000138863 |
fam173b
|
family with sequence similarity 173, member B |
| chr19_+_48049202 | 0.33 |
ENSDART00000027158
|
psmd3
|
proteasome 26S subunit, non-ATPase 3 |
| chr13_+_15701849 | 0.32 |
ENSDART00000003517
|
trmt61a
|
tRNA methyltransferase 61A |
| chr5_-_23179319 | 0.32 |
ENSDART00000161883
ENSDART00000136260 |
si:dkey-114c15.5
|
si:dkey-114c15.5 |
| chr22_+_336256 | 0.31 |
ENSDART00000019155
|
btg2
|
B-cell translocation gene 2 |
| chr11_-_1509773 | 0.31 |
ENSDART00000050762
|
phactr3b
|
phosphatase and actin regulator 3b |
| chr12_-_5728755 | 0.31 |
ENSDART00000105887
|
dlx4b
|
distal-less homeobox 4b |
| chr3_+_14611299 | 0.30 |
ENSDART00000140577
|
tspan35
|
tetraspanin 35 |
| chr16_+_33655890 | 0.30 |
ENSDART00000143757
|
fhl3a
|
four and a half LIM domains 3a |
| chr16_-_7379328 | 0.30 |
ENSDART00000060447
|
med18
|
mediator of RNA polymerase II transcription, subunit 18 homolog (yeast) |
| chr8_+_21159122 | 0.30 |
ENSDART00000033491
|
spryd4
|
SPRY domain containing 4 |
| chr14_+_38786298 | 0.30 |
ENSDART00000164440
|
si:ch211-195b11.3
|
si:ch211-195b11.3 |
| chr9_+_30108641 | 0.30 |
ENSDART00000060174
|
jagn1a
|
jagunal homolog 1a |
| chr8_+_23725957 | 0.30 |
ENSDART00000104346
|
mkrn4
|
makorin, ring finger protein, 4 |
| chr14_+_16034447 | 0.30 |
ENSDART00000161348
|
prelid1a
|
PRELI domain containing 1a |
| chr7_+_44715224 | 0.30 |
ENSDART00000184630
|
si:dkey-56m19.5
|
si:dkey-56m19.5 |
| chr4_+_5249494 | 0.29 |
ENSDART00000150391
|
si:ch211-214j24.14
|
si:ch211-214j24.14 |
| chr6_-_10912424 | 0.29 |
ENSDART00000036456
|
cycsb
|
cytochrome c, somatic b |
| chr8_-_40327397 | 0.29 |
ENSDART00000074125
|
aplnra
|
apelin receptor a |
| chr13_-_45022527 | 0.29 |
ENSDART00000159021
|
khdrbs1a
|
KH domain containing, RNA binding, signal transduction associated 1a |
| chr5_-_30615901 | 0.29 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr14_-_17588345 | 0.29 |
ENSDART00000143486
|
selenot2
|
selenoprotein T, 2 |
| chr19_+_15440841 | 0.29 |
ENSDART00000182329
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr15_-_18177124 | 0.29 |
ENSDART00000156505
|
tmprss5
|
transmembrane protease, serine 5 |
| chr16_-_12060488 | 0.29 |
ENSDART00000188733
|
si:ch211-69g19.2
|
si:ch211-69g19.2 |
| chr15_-_20528494 | 0.29 |
ENSDART00000048423
|
timm50
|
translocase of inner mitochondrial membrane 50 homolog (S. cerevisiae) |
| chr7_-_74090168 | 0.28 |
ENSDART00000050528
|
tyrp1a
|
tyrosinase-related protein 1a |
| chr2_+_12255568 | 0.28 |
ENSDART00000184164
ENSDART00000013454 |
prtfdc1
|
phosphoribosyl transferase domain containing 1 |
| chr2_-_51096647 | 0.28 |
ENSDART00000167172
|
THEM6
|
si:ch73-52e5.2 |
| chr3_+_39568290 | 0.28 |
ENSDART00000020741
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr2_+_15048410 | 0.28 |
ENSDART00000058484
|
cnn3b
|
calponin 3, acidic b |
| chr23_+_9560797 | 0.27 |
ENSDART00000180014
|
adrm1
|
adhesion regulating molecule 1 |
| chr25_-_12822239 | 0.27 |
ENSDART00000166411
|
uba2
|
ubiquitin-like modifier activating enzyme 2 |
| chr10_-_27009413 | 0.27 |
ENSDART00000139942
ENSDART00000146983 ENSDART00000132352 |
uqcc3
|
ubiquinol-cytochrome c reductase complex assembly factor 3 |
| chr3_+_30500968 | 0.27 |
ENSDART00000103447
|
si:dkey-13n23.3
|
si:dkey-13n23.3 |
| chr17_+_38262408 | 0.27 |
ENSDART00000017493
|
nkx2.1
|
NK2 homeobox 1 |
| chr23_-_12345764 | 0.27 |
ENSDART00000133956
|
phactr3a
|
phosphatase and actin regulator 3a |
| chr2_+_23790748 | 0.27 |
ENSDART00000041877
|
csrnp1a
|
cysteine-serine-rich nuclear protein 1a |
| chr13_-_31647323 | 0.27 |
ENSDART00000135381
|
six4a
|
SIX homeobox 4a |
| chr22_-_21176269 | 0.26 |
ENSDART00000112839
|
rex1bd
|
required for excision 1-B domain containing |
| chr3_-_15487111 | 0.26 |
ENSDART00000011320
|
nfatc2ip
|
nuclear factor of activated T cells 2 interacting protein |
| chr20_+_28364742 | 0.26 |
ENSDART00000103355
|
rhov
|
ras homolog family member V |
| chr1_+_17695426 | 0.26 |
ENSDART00000103236
|
ankrd37
|
ankyrin repeat domain 37 |
| chr23_-_27589508 | 0.26 |
ENSDART00000178404
|
si:ch211-156j22.4
|
si:ch211-156j22.4 |
| chr11_+_3254252 | 0.26 |
ENSDART00000123568
|
pmela
|
premelanosome protein a |
| chr3_+_27027781 | 0.26 |
ENSDART00000065495
|
emp2
|
epithelial membrane protein 2 |
| chr1_-_30979707 | 0.26 |
ENSDART00000008469
|
dlx2b
|
distal-less homeobox 2b |
| chr10_-_34033680 | 0.26 |
ENSDART00000180734
|
rfc3
|
replication factor C (activator 1) 3 |
| chr2_-_15040345 | 0.26 |
ENSDART00000109657
|
si:dkey-10f21.4
|
si:dkey-10f21.4 |
| chr23_-_18805567 | 0.26 |
ENSDART00000143065
ENSDART00000190245 |
nsfl1c
|
NSFL1 (p97) cofactor (p47) |
| chr16_-_45069882 | 0.25 |
ENSDART00000058384
|
gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr2_+_25378457 | 0.25 |
ENSDART00000089108
|
fndc3ba
|
fibronectin type III domain containing 3Ba |
| chr3_-_23604396 | 0.25 |
ENSDART00000078423
|
atp5mc1
|
ATP synthase membrane subunit c locus 1 |
| chr20_+_30378803 | 0.25 |
ENSDART00000148242
ENSDART00000169140 ENSDART00000062441 |
rnaseh1
|
ribonuclease H1 |
| chr24_-_21903360 | 0.25 |
ENSDART00000091252
|
spata13
|
spermatogenesis associated 13 |
| chr22_-_17677947 | 0.25 |
ENSDART00000139911
|
tjp3
|
tight junction protein 3 |
| chr24_-_4148914 | 0.25 |
ENSDART00000033494
ENSDART00000147849 |
klf6a
|
Kruppel-like factor 6a |
| chr8_+_21353878 | 0.25 |
ENSDART00000056420
|
alas2
|
aminolevulinate, delta-, synthase 2 |
| chr13_-_28688104 | 0.25 |
ENSDART00000133827
|
pcgf6
|
polycomb group ring finger 6 |
| chr4_+_38981587 | 0.25 |
ENSDART00000142713
|
si:dkey-66k12.3
|
si:dkey-66k12.3 |
| chr17_-_21784152 | 0.25 |
ENSDART00000127254
|
hmx2
|
H6 family homeobox 2 |
| chr20_+_400487 | 0.25 |
ENSDART00000108872
|
ndufaf4
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 4 |
| chr12_-_48960308 | 0.25 |
ENSDART00000176247
|
CABZ01112647.1
|
|
| chr5_+_57320113 | 0.25 |
ENSDART00000036331
|
atp6v1g1
|
ATPase H+ transporting V1 subunit G1 |
| chr24_+_23730061 | 0.25 |
ENSDART00000080343
|
ppp1r42
|
protein phosphatase 1, regulatory subunit 42 |
| chr16_-_47301376 | 0.25 |
ENSDART00000169697
|
mios
|
missing oocyte, meiosis regulator, homolog (Drosophila) |
| chr18_+_45645357 | 0.24 |
ENSDART00000010256
|
eif3m
|
eukaryotic translation initiation factor 3, subunit M |
| chr4_-_5302866 | 0.24 |
ENSDART00000138590
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr4_-_965267 | 0.24 |
ENSDART00000093289
|
atp23
|
ATP23 metallopeptidase and ATP synthase assembly factor homolog |
| chr5_+_42064144 | 0.24 |
ENSDART00000035235
|
si:ch211-202a12.4
|
si:ch211-202a12.4 |
| chr1_-_14506759 | 0.24 |
ENSDART00000057044
|
si:dkey-194g4.1
|
si:dkey-194g4.1 |
| chr24_-_21903588 | 0.24 |
ENSDART00000180991
|
spata13
|
spermatogenesis associated 13 |
| chr5_+_4533244 | 0.24 |
ENSDART00000158826
|
CABZ01058650.1
|
Danio rerio thiosulfate sulfurtransferase/rhodanese-like domain-containing protein 1 (LOC561325), mRNA. |
| chr4_+_6639292 | 0.24 |
ENSDART00000158573
|
SMIM30
|
si:dkey-112e7.2 |
| chr11_-_497854 | 0.24 |
ENSDART00000104520
|
cnbpb
|
CCHC-type zinc finger, nucleic acid binding protein b |
| chr7_+_15871156 | 0.24 |
ENSDART00000145946
|
pax6b
|
paired box 6b |
| chr3_+_58092212 | 0.24 |
ENSDART00000156059
|
si:ch211-256e16.7
|
si:ch211-256e16.7 |
| chr24_-_35534273 | 0.24 |
ENSDART00000026578
|
ube2v2
|
ubiquitin-conjugating enzyme E2 variant 2 |
| chr5_+_34997763 | 0.24 |
ENSDART00000043341
|
foxd1
|
forkhead box D1 |
| chr6_-_46589726 | 0.24 |
ENSDART00000084334
|
ptgis
|
prostaglandin I2 (prostacyclin) synthase |
| chr8_-_18203092 | 0.23 |
ENSDART00000140620
|
elovl8b
|
ELOVL fatty acid elongase 8b |
| chr21_+_45816030 | 0.23 |
ENSDART00000187056
|
pitx1
|
paired-like homeodomain 1 |
| chr5_-_67365333 | 0.23 |
ENSDART00000133438
|
unga
|
uracil DNA glycosylase a |
| chr3_+_31621774 | 0.23 |
ENSDART00000076636
|
fzd2
|
frizzled class receptor 2 |
| chr19_-_17864213 | 0.23 |
ENSDART00000151043
|
ints8
|
integrator complex subunit 8 |
| chr7_-_28647959 | 0.23 |
ENSDART00000150148
|
slc7a6
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 6 |
| chr7_+_42206543 | 0.23 |
ENSDART00000112543
|
phkb
|
phosphorylase kinase, beta |
| chr16_+_23960933 | 0.23 |
ENSDART00000146077
|
apoeb
|
apolipoprotein Eb |
| chr18_+_40381102 | 0.23 |
ENSDART00000136588
|
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr2_-_37896965 | 0.23 |
ENSDART00000129852
|
hbl1
|
hexose-binding lectin 1 |
| chr22_+_16759010 | 0.23 |
ENSDART00000079638
ENSDART00000113099 |
tm2d1
|
TM2 domain containing 1 |
| chr16_+_23960744 | 0.23 |
ENSDART00000058965
|
apoeb
|
apolipoprotein Eb |
| chr12_-_31484677 | 0.23 |
ENSDART00000066578
|
tectb
|
tectorin beta |
| chr8_-_17790916 | 0.23 |
ENSDART00000189020
ENSDART00000131276 |
si:ch211-150o23.2
|
si:ch211-150o23.2 |
| chr1_+_16463079 | 0.23 |
ENSDART00000078675
|
vps37a
|
vacuolar protein sorting 37A |
| chr22_-_21676364 | 0.23 |
ENSDART00000183668
|
tle2b
|
transducin like enhancer of split 2b |
| chr7_-_35516251 | 0.23 |
ENSDART00000045628
|
irx6a
|
iroquois homeobox 6a |
| chr17_-_43022468 | 0.23 |
ENSDART00000002945
|
NPC2 (1 of many)
|
zgc:193725 |
| chr14_+_34547554 | 0.23 |
ENSDART00000074819
|
gabrp
|
gamma-aminobutyric acid (GABA) A receptor, pi |
| chr15_-_23342752 | 0.23 |
ENSDART00000020425
|
mcamb
|
melanoma cell adhesion molecule b |
| chr14_-_24410673 | 0.23 |
ENSDART00000125923
|
cxcl14
|
chemokine (C-X-C motif) ligand 14 |
| chr3_-_15475067 | 0.23 |
ENSDART00000025324
ENSDART00000139575 |
spns1
|
spinster homolog 1 (Drosophila) |
| chr23_+_2421689 | 0.23 |
ENSDART00000180200
|
tcp1
|
t-complex 1 |
| chr25_-_19608382 | 0.23 |
ENSDART00000022279
ENSDART00000135201 ENSDART00000147223 ENSDART00000190220 ENSDART00000184242 ENSDART00000166824 |
gtse1
|
G-2 and S-phase expressed 1 |
| chr10_+_38560032 | 0.23 |
ENSDART00000177197
|
LO018336.1
|
|
| chr1_+_51475094 | 0.23 |
ENSDART00000146352
|
meis1a
|
Meis homeobox 1 a |
| chr10_-_34033455 | 0.23 |
ENSDART00000131685
|
rfc3
|
replication factor C (activator 1) 3 |
| chr18_-_7137153 | 0.22 |
ENSDART00000019571
|
cd9a
|
CD9 molecule a |
| chr2_-_58257624 | 0.22 |
ENSDART00000098940
|
foxl2b
|
forkhead box L2b |
| chr12_+_10163585 | 0.22 |
ENSDART00000106191
|
psmc5
|
proteasome 26S subunit, ATPase 5 |
| chr24_+_31334209 | 0.22 |
ENSDART00000168837
ENSDART00000172473 |
fam168b
|
family with sequence similarity 168, member B |
| chr7_+_24390939 | 0.22 |
ENSDART00000087494
ENSDART00000125463 |
haus3
|
HAUS augmin-like complex, subunit 3 |
| chr18_-_3552414 | 0.22 |
ENSDART00000163762
ENSDART00000165434 ENSDART00000161197 ENSDART00000166841 ENSDART00000170260 |
dcun1d5
|
DCN1, defective in cullin neddylation 1, domain containing 5 (S. cerevisiae) |
| chr20_-_28842524 | 0.22 |
ENSDART00000046035
ENSDART00000139843 ENSDART00000129858 ENSDART00000137425 ENSDART00000135720 |
max
|
myc associated factor X |
| chr25_+_1591964 | 0.22 |
ENSDART00000093277
|
ppm1h
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H |
| chr5_+_4298636 | 0.22 |
ENSDART00000100061
|
prdx4
|
peroxiredoxin 4 |
| chr3_-_4501026 | 0.22 |
ENSDART00000163052
|
zgc:162198
|
zgc:162198 |
| chr25_-_17378881 | 0.22 |
ENSDART00000064586
|
cyp2x7
|
cytochrome P450, family 2, subfamily X, polypeptide 7 |
| chr8_-_14091886 | 0.22 |
ENSDART00000137857
|
si:ch211-229n2.7
|
si:ch211-229n2.7 |
| chr5_-_22619879 | 0.22 |
ENSDART00000051623
|
zgc:113208
|
zgc:113208 |
| chr18_+_44567191 | 0.22 |
ENSDART00000086943
|
dhx34
|
DEAH (Asp-Glu-Ala-His) box polypeptide 34 |
| chr19_+_2546775 | 0.21 |
ENSDART00000148527
ENSDART00000097528 |
sp4
|
sp4 transcription factor |
| chr8_+_17167876 | 0.21 |
ENSDART00000134665
|
cenph
|
centromere protein H |
| chr8_-_30944465 | 0.21 |
ENSDART00000128792
ENSDART00000191717 ENSDART00000049944 |
smarcb1a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1a |
| chr8_+_11471350 | 0.21 |
ENSDART00000092355
ENSDART00000136184 |
tjp2b
|
tight junction protein 2b (zona occludens 2) |
| chr13_+_421231 | 0.21 |
ENSDART00000188212
ENSDART00000017854 |
lgi1a
|
leucine-rich, glioma inactivated 1a |
| chr15_+_46853505 | 0.21 |
ENSDART00000159844
|
zgc:153039
|
zgc:153039 |
| chr13_-_36535128 | 0.21 |
ENSDART00000043312
|
srsf5a
|
serine/arginine-rich splicing factor 5a |
| chr22_+_22438783 | 0.21 |
ENSDART00000147825
|
kif14
|
kinesin family member 14 |
| chr3_-_20091964 | 0.21 |
ENSDART00000029386
ENSDART00000020253 ENSDART00000124326 |
slc4a1a
|
solute carrier family 4 (anion exchanger), member 1a (Diego blood group) |
| chr7_+_29992889 | 0.21 |
ENSDART00000055936
|
isl2b
|
ISL LIM homeobox 2b |
| chr13_-_9841806 | 0.21 |
ENSDART00000101949
|
sfxn4
|
sideroflexin 4 |
| chr24_-_39186185 | 0.21 |
ENSDART00000123019
ENSDART00000191114 |
nubp2
|
nucleotide binding protein 2 (MinD homolog, E. coli) |
| chr18_-_15911394 | 0.21 |
ENSDART00000091339
|
plekhg7
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 7 |
| chr9_+_426392 | 0.21 |
ENSDART00000172515
|
bzw1b
|
basic leucine zipper and W2 domains 1b |
| chr23_+_19675780 | 0.21 |
ENSDART00000124335
|
smim4
|
small integral membrane protein 4 |
| chr16_+_28728347 | 0.21 |
ENSDART00000149240
|
si:dkey-24i24.3
|
si:dkey-24i24.3 |
| chr5_+_67971627 | 0.21 |
ENSDART00000144879
|
mtif3
|
mitochondrial translational initiation factor 3 |
| chr8_-_44904723 | 0.21 |
ENSDART00000040804
|
praf2
|
PRA1 domain family, member 2 |
| chr3_-_21106093 | 0.21 |
ENSDART00000156566
|
maza
|
MYC-associated zinc finger protein a (purine-binding transcription factor) |
| chr2_+_11685742 | 0.21 |
ENSDART00000138562
|
greb1l
|
growth regulation by estrogen in breast cancer-like |
| chr4_-_49952636 | 0.21 |
ENSDART00000157941
|
si:dkey-156k2.3
|
si:dkey-156k2.3 |
| chr4_-_9780931 | 0.20 |
ENSDART00000134280
ENSDART00000150664 ENSDART00000150304 ENSDART00000080744 |
svopl
|
SVOP-like |
| chr4_-_4756349 | 0.20 |
ENSDART00000057507
|
ssbp1
|
single-stranded DNA binding protein 1 |
| chr21_-_39604549 | 0.20 |
ENSDART00000161516
ENSDART00000151638 |
pigs
|
phosphatidylinositol glycan anchor biosynthesis, class S |
| chr22_-_5744491 | 0.20 |
ENSDART00000145014
|
bcdin3d
|
BCDIN3 domain containing |
| chr14_+_26719691 | 0.20 |
ENSDART00000078522
ENSDART00000172927 |
eef1g
|
eukaryotic translation elongation factor 1 gamma |
| chr14_-_6936731 | 0.20 |
ENSDART00000162196
|
clk4a
|
CDC-like kinase 4a |
| chr22_-_26852516 | 0.20 |
ENSDART00000005829
|
gde1
|
glycerophosphodiester phosphodiesterase 1 |
| chr4_-_70488123 | 0.20 |
ENSDART00000169266
|
si:dkeyp-44b5.5
|
si:dkeyp-44b5.5 |
| chr24_-_33756003 | 0.20 |
ENSDART00000079283
|
tmeff1b
|
transmembrane protein with EGF-like and two follistatin-like domains 1b |
| chr8_+_23639124 | 0.20 |
ENSDART00000083108
|
nt5dc2
|
5'-nucleotidase domain containing 2 |
| chr9_+_21819082 | 0.20 |
ENSDART00000136902
ENSDART00000101991 |
txndc9
|
thioredoxin domain containing 9 |
| chr15_-_41245962 | 0.20 |
ENSDART00000155359
|
smco4
|
single-pass membrane protein with coiled-coil domains 4 |
| chr13_+_35925490 | 0.20 |
ENSDART00000046115
|
mfsd2aa
|
major facilitator superfamily domain containing 2aa |
| chr10_-_23099809 | 0.20 |
ENSDART00000148333
ENSDART00000079703 ENSDART00000162444 |
nle1
|
notchless homolog 1 (Drosophila) |
| chr5_-_31689796 | 0.20 |
ENSDART00000184319
ENSDART00000190229 ENSDART00000186294 ENSDART00000170313 ENSDART00000147065 ENSDART00000134427 ENSDART00000098172 |
sh3glb2b
|
SH3-domain GRB2-like endophilin B2b |
| chr7_+_27455321 | 0.20 |
ENSDART00000148417
|
cyp2r1
|
cytochrome P450, family 2, subfamily R, polypeptide 1 |
| chr19_+_37120491 | 0.20 |
ENSDART00000032341
|
pef1
|
penta-EF-hand domain containing 1 |
| chr17_+_51224421 | 0.20 |
ENSDART00000025229
|
adi1
|
acireductone dioxygenase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of pax2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0007585 | respiratory gaseous exchange(GO:0007585) regulation of respiratory gaseous exchange(GO:0043576) |
| 0.2 | 1.7 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.2 | 0.5 | GO:0071830 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.1 | 0.6 | GO:0097510 | base-excision repair, AP site formation via deaminated base removal(GO:0097510) |
| 0.1 | 0.3 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.1 | 0.3 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.1 | 0.3 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 0.1 | 0.4 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.1 | 0.3 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.4 | GO:0006843 | mitochondrial citrate transport(GO:0006843) |
| 0.1 | 0.3 | GO:0097435 | fibril organization(GO:0097435) |
| 0.1 | 0.3 | GO:0003071 | renal system process involved in regulation of blood volume(GO:0001977) renal system process involved in regulation of systemic arterial blood pressure(GO:0003071) regulation of glomerular filtration(GO:0003093) |
| 0.1 | 0.3 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 0.1 | 0.5 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 0.2 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.1 | 0.2 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) |
| 0.1 | 0.2 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 0.3 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903589) |
| 0.1 | 0.4 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.1 | 0.5 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 0.2 | GO:0051095 | regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.1 | 0.4 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.1 | 0.2 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.1 | 0.3 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.1 | 0.2 | GO:0060571 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 0.1 | 0.2 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.1 | 0.6 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.1 | 0.2 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 0.1 | 0.2 | GO:1902373 | negative regulation of mRNA catabolic process(GO:1902373) regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 0.2 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.3 | GO:0021570 | rhombomere 4 development(GO:0021570) rhombomere 4 morphogenesis(GO:0021661) |
| 0.0 | 0.2 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.0 | 0.2 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.0 | 0.3 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.2 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.0 | 0.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.7 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.0 | 0.1 | GO:1903334 | regulation of protein folding(GO:1903332) positive regulation of protein folding(GO:1903334) regulation of chaperone-mediated protein folding(GO:1903644) positive regulation of chaperone-mediated protein folding(GO:1903646) |
| 0.0 | 0.2 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.0 | 0.1 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.2 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.2 | GO:0070292 | N-acylphosphatidylethanolamine metabolic process(GO:0070292) |
| 0.0 | 0.3 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.0 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 0.3 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.2 | GO:1903723 | negative regulation of centriole elongation(GO:1903723) |
| 0.0 | 0.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.3 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.4 | GO:0001836 | release of cytochrome c from mitochondria(GO:0001836) |
| 0.0 | 0.3 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.7 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.2 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.0 | 0.2 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.0 | 0.2 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 0.2 | GO:0071265 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.0 | 0.2 | GO:0035989 | tendon development(GO:0035989) |
| 0.0 | 0.2 | GO:0071480 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.1 | GO:0046443 | FAD biosynthetic process(GO:0006747) FAD metabolic process(GO:0046443) flavin adenine dinucleotide metabolic process(GO:0072387) flavin adenine dinucleotide biosynthetic process(GO:0072388) |
| 0.0 | 0.2 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.1 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.0 | 0.4 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.2 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.1 | GO:1901255 | nucleotide-excision repair involved in interstrand cross-link repair(GO:1901255) |
| 0.0 | 0.1 | GO:0021543 | pallium development(GO:0021543) cerebral cortex cell migration(GO:0021795) cerebral cortex tangential migration(GO:0021800) cerebral cortex tangential migration using cell-axon interactions(GO:0021824) substrate-dependent cerebral cortex tangential migration(GO:0021825) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) cerebral cortex development(GO:0021987) |
| 0.0 | 0.1 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 0.0 | 0.2 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.0 | 0.6 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.2 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.2 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.0 | 0.2 | GO:1901166 | neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.0 | 0.2 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.1 | GO:0071514 | genetic imprinting(GO:0071514) |
| 0.0 | 0.2 | GO:0071295 | cellular response to nutrient(GO:0031670) vitamin D3 metabolic process(GO:0070640) cellular response to vitamin(GO:0071295) cellular response to vitamin D(GO:0071305) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.3 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.3 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.0 | 0.6 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.2 | GO:0006083 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.0 | 0.2 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.1 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.0 | 0.1 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.1 | GO:0036314 | response to sterol(GO:0036314) cellular response to sterol(GO:0036315) SREBP-SCAP complex retention in endoplasmic reticulum(GO:0036316) regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.1 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 0.0 | 0.3 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.1 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.0 | 0.6 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.1 | GO:0015729 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.0 | 0.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.2 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.0 | 0.1 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.2 | GO:0002312 | B cell activation involved in immune response(GO:0002312) isotype switching(GO:0045190) |
| 0.0 | 0.1 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.0 | 0.1 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.4 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.2 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.0 | 0.3 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.0 | 0.2 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.2 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.1 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.0 | 0.7 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.7 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.1 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 0.0 | 0.1 | GO:0072091 | forebrain anterior/posterior pattern specification(GO:0021797) regulation of stem cell proliferation(GO:0072091) |
| 0.0 | 0.1 | GO:0042762 | regulation of sulfur metabolic process(GO:0042762) |
| 0.0 | 0.1 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.1 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.0 | 0.1 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.0 | 0.1 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 0.1 | GO:1904667 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.0 | 0.1 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.2 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.0 | 0.1 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 | 0.4 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.2 | GO:0030719 | P granule organization(GO:0030719) |
| 0.0 | 0.1 | GO:0010719 | photoreceptor cell morphogenesis(GO:0008594) negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.0 | 0.6 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.1 | GO:0046103 | adenosine catabolic process(GO:0006154) inosine metabolic process(GO:0046102) inosine biosynthetic process(GO:0046103) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.1 | GO:0016037 | light absorption(GO:0016037) |
| 0.0 | 0.2 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.5 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.1 | GO:0045905 | positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:1903523 | negative regulation of heart contraction(GO:0045822) negative regulation of blood circulation(GO:1903523) |
| 0.0 | 0.2 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.2 | GO:0016576 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.5 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.1 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 0.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.2 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 0.0 | 0.1 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.1 | GO:0006551 | leucine metabolic process(GO:0006551) leucine catabolic process(GO:0006552) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.1 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.2 | GO:0001516 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 | 0.0 | GO:0045621 | positive regulation of lymphocyte differentiation(GO:0045621) |
| 0.0 | 0.1 | GO:0034695 | response to prostaglandin E(GO:0034695) cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.0 | 0.1 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.0 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.1 | GO:0048662 | regulation of smooth muscle cell proliferation(GO:0048660) negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.0 | 0.3 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 0.2 | GO:0051443 | protein neddylation(GO:0045116) regulation of ubiquitin-protein transferase activity(GO:0051438) positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 | 0.1 | GO:0070293 | renal absorption(GO:0070293) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.3 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.6 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.2 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.2 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.1 | GO:0015740 | C4-dicarboxylate transport(GO:0015740) acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.4 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.1 | GO:0090024 | negative regulation of leukocyte chemotaxis(GO:0002689) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.0 | 0.3 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.2 | GO:0006999 | NLS-bearing protein import into nucleus(GO:0006607) nuclear pore organization(GO:0006999) |
| 0.0 | 0.1 | GO:0048823 | nucleate erythrocyte development(GO:0048823) |
| 0.0 | 0.4 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.3 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.2 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.1 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.1 | GO:0043393 | regulation of protein binding(GO:0043393) |
| 0.0 | 0.3 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.1 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.0 | 0.2 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 0.1 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 0.2 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.0 | 0.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0021744 | medulla oblongata development(GO:0021550) dorsal motor nucleus of vagus nerve development(GO:0021744) |
| 0.0 | 0.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.2 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.1 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.4 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.1 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 0.1 | GO:1903725 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) regulation of phospholipid metabolic process(GO:1903725) |
| 0.0 | 0.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.3 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.0 | 0.2 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.1 | GO:0046473 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.1 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.0 | GO:0018315 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.0 | 0.1 | GO:0046337 | phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.0 | 0.5 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.1 | 1.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.3 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 0.3 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 0.5 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 0.3 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.1 | 0.3 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.1 | 0.5 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 0.2 | GO:0097541 | axonemal basal plate(GO:0097541) |
| 0.1 | 0.2 | GO:0042721 | mitochondrial intermembrane space protein transporter complex(GO:0042719) mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.1 | GO:0072380 | TRC complex(GO:0072380) |
| 0.0 | 0.1 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.2 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.2 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.3 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.1 | GO:1990879 | CST complex(GO:1990879) |
| 0.0 | 0.3 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.3 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.2 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.2 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.2 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.2 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.1 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.0 | 0.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.4 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.2 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.2 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.2 | GO:0035060 | brahma complex(GO:0035060) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.1 | GO:0090443 | FAR/SIN/STRIPAK complex(GO:0090443) |
| 0.0 | 0.3 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.1 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.0 | 0.1 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.0 | 0.1 | GO:0016234 | inclusion body(GO:0016234) Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.1 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.1 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 0.0 | 0.2 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.4 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.1 | GO:0008247 | 1-alkyl-2-acetylglycerophosphocholine esterase complex(GO:0008247) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.1 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 0.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.0 | 0.5 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.3 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.1 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.2 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.0 | 0.1 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 0.2 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.2 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.0 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.2 | GO:0033176 | proton-transporting V-type ATPase complex(GO:0033176) |
| 0.0 | 0.1 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.0 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.0 | 0.2 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.2 | 0.6 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.2 | 0.5 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 0.8 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.3 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.1 | 0.3 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 0.3 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.1 | 0.2 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.1 | 0.7 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.4 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.1 | 0.2 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.1 | 0.3 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.1 | 0.3 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.1 | 0.3 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.1 | 0.3 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.1 | 0.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.3 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.2 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.1 | 0.2 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 0.2 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.1 | 0.3 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.1 | 0.4 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 0.2 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.1 | 0.2 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.1 | 0.4 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 0.2 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.0 | 0.2 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.0 | 0.2 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.4 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.4 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.2 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.5 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.2 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.3 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.8 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.1 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.0 | 0.1 | GO:0003919 | FMN adenylyltransferase activity(GO:0003919) |
| 0.0 | 0.3 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.2 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.1 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.1 | GO:0047690 | aspartyltransferase activity(GO:0047690) |
| 0.0 | 0.1 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.0 | 0.1 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.0 | 0.3 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.6 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.2 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 0.1 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.5 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.0 | 0.1 | GO:0015117 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.2 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.3 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.4 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.2 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.2 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.1 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.0 | 0.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 0.2 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.1 | GO:0097001 | sphingolipid binding(GO:0046625) ceramide binding(GO:0097001) |
| 0.0 | 0.1 | GO:0043394 | glycoprotein binding(GO:0001948) proteoglycan binding(GO:0043394) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.2 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.2 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.1 | GO:0043734 | DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.1 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 0.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.3 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.1 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.6 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0016521 | pituitary adenylate cyclase activating polypeptide activity(GO:0016521) |
| 0.0 | 0.2 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.1 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.1 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.2 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.1 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.2 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.5 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.1 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.2 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.1 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 1.0 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.6 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 0.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.3 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.0 | 0.6 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.0 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.0 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.0 | 0.0 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.0 | 0.2 | GO:0016860 | intramolecular oxidoreductase activity(GO:0016860) |
| 0.0 | 0.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.3 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.9 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.5 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.5 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.1 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.2 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.2 | PID MYC PATHWAY | C-MYC pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 0.6 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.8 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 1.5 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 1.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.2 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.6 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.4 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.6 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.6 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.5 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.3 | REACTOME ORC1 REMOVAL FROM CHROMATIN | Genes involved in Orc1 removal from chromatin |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.4 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 0.1 | REACTOME CDT1 ASSOCIATION WITH THE CDC6 ORC ORIGIN COMPLEX | Genes involved in CDT1 association with the CDC6:ORC:origin complex |
| 0.0 | 0.4 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.5 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.7 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.2 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.0 | 0.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.2 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |