Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
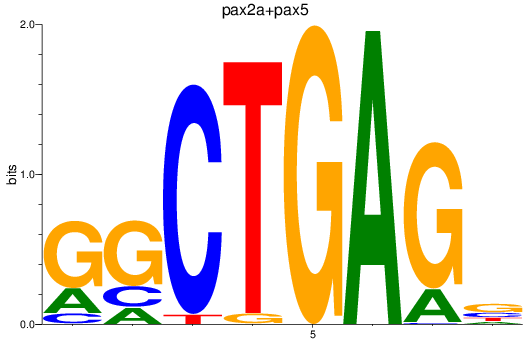
Results for pax2a+pax5
Z-value: 1.26
Transcription factors associated with pax2a+pax5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pax2a
|
ENSDARG00000028148 | paired box 2a |
|
pax5
|
ENSDARG00000037383 | paired box 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pax2a | dr11_v1_chr13_+_29771463_29771463 | 0.67 | 1.5e-03 | Click! |
| pax5 | dr11_v1_chr1_+_21725821_21725821 | -0.49 | 3.5e-02 | Click! |
Activity profile of pax2a+pax5 motif
Sorted Z-values of pax2a+pax5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_22318511 | 2.45 |
ENSDART00000129295
|
crygm2d2
|
crystallin, gamma M2d2 |
| chr12_+_13256415 | 2.33 |
ENSDART00000144542
|
atp2a1l
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1, like |
| chr3_-_61205711 | 2.29 |
ENSDART00000055062
|
pvalb1
|
parvalbumin 1 |
| chr1_+_14283692 | 2.00 |
ENSDART00000017679
|
ppp2r2ca
|
protein phosphatase 2, regulatory subunit B, gamma a |
| chr11_-_44543082 | 1.91 |
ENSDART00000099568
|
gpr137bb
|
G protein-coupled receptor 137Bb |
| chr7_+_29952169 | 1.83 |
ENSDART00000173540
ENSDART00000173940 ENSDART00000173906 ENSDART00000173772 ENSDART00000173506 ENSDART00000039657 |
tpma
|
alpha-tropomyosin |
| chr24_-_41312459 | 1.78 |
ENSDART00000041349
|
crygn2
|
crystallin, gamma N2 |
| chr9_-_22232902 | 1.71 |
ENSDART00000101845
|
crygm2d5
|
crystallin, gamma M2d5 |
| chr5_+_36974931 | 1.66 |
ENSDART00000193063
|
gjd1a
|
gap junction protein delta 1a |
| chr2_+_32780138 | 1.51 |
ENSDART00000082250
|
zgc:136930
|
zgc:136930 |
| chr19_-_103289 | 1.50 |
ENSDART00000143118
|
adgrb1b
|
adhesion G protein-coupled receptor B1b |
| chr7_+_29951997 | 1.45 |
ENSDART00000173453
|
tpma
|
alpha-tropomyosin |
| chr5_-_57641257 | 1.42 |
ENSDART00000149282
|
hspb2
|
heat shock protein, alpha-crystallin-related, b2 |
| chr3_+_32526799 | 1.41 |
ENSDART00000185755
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr25_-_225964 | 1.39 |
ENSDART00000193424
|
CABZ01113818.1
|
|
| chr20_-_17041025 | 1.38 |
ENSDART00000063764
|
si:dkey-5n18.1
|
si:dkey-5n18.1 |
| chr8_-_23416362 | 1.32 |
ENSDART00000063005
|
gpr173
|
G protein-coupled receptor 173 |
| chr16_-_13730152 | 1.31 |
ENSDART00000138772
|
ttyh1
|
tweety family member 1 |
| chr7_+_10610791 | 1.30 |
ENSDART00000166064
|
fah
|
fumarylacetoacetate hydrolase (fumarylacetoacetase) |
| chr9_-_42989297 | 1.24 |
ENSDART00000126871
|
ttn.2
|
titin, tandem duplicate 2 |
| chr14_-_36378494 | 1.22 |
ENSDART00000058503
|
gpm6aa
|
glycoprotein M6Aa |
| chr12_-_35787801 | 1.20 |
ENSDART00000171682
|
aatkb
|
apoptosis-associated tyrosine kinase b |
| chr25_+_6306885 | 1.13 |
ENSDART00000142705
ENSDART00000067510 |
crabp1a
|
cellular retinoic acid binding protein 1a |
| chr16_-_560574 | 1.12 |
ENSDART00000148452
|
irx2a
|
iroquois homeobox 2a |
| chr24_-_2947393 | 1.09 |
ENSDART00000166661
ENSDART00000147110 |
tubb6
|
tubulin, beta 6 class V |
| chr9_-_12034444 | 1.08 |
ENSDART00000038651
|
znf804a
|
zinc finger protein 804A |
| chr18_+_22302635 | 1.08 |
ENSDART00000141051
|
carmil2
|
capping protein regulator and myosin 1 linker 2 |
| chr6_-_14139503 | 1.05 |
ENSDART00000089577
|
cacnb4b
|
calcium channel, voltage-dependent, beta 4b subunit |
| chr8_-_46897734 | 1.04 |
ENSDART00000138125
|
hes2.2
|
hes family bHLH transcription factor 2, tandem duplicate 2 |
| chr14_+_46313396 | 1.04 |
ENSDART00000047525
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr21_+_5209716 | 1.02 |
ENSDART00000102539
ENSDART00000053148 ENSDART00000102536 |
st8sia5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5 |
| chr24_+_42948 | 1.02 |
ENSDART00000122785
|
tmx3b
|
thioredoxin related transmembrane protein 3b |
| chr18_+_9635178 | 1.01 |
ENSDART00000183486
|
pclob
|
piccolo presynaptic cytomatrix protein b |
| chr3_-_28250722 | 1.00 |
ENSDART00000165936
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr1_-_29045426 | 1.00 |
ENSDART00000019770
|
gpm6ba
|
glycoprotein M6Ba |
| chr7_+_29952719 | 1.00 |
ENSDART00000173737
|
tpma
|
alpha-tropomyosin |
| chr10_+_11309814 | 0.99 |
ENSDART00000145673
|
si:ch211-126i22.5
|
si:ch211-126i22.5 |
| chr3_+_17806213 | 0.98 |
ENSDART00000055890
|
znf385c
|
zinc finger protein 385C |
| chr19_-_48019366 | 0.98 |
ENSDART00000169429
|
si:ch1073-205c8.5
|
si:ch1073-205c8.5 |
| chr24_+_3963684 | 0.98 |
ENSDART00000182959
ENSDART00000185926 ENSDART00000167043 ENSDART00000033394 |
pfkpa
|
phosphofructokinase, platelet a |
| chr3_-_28258462 | 0.97 |
ENSDART00000191573
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr16_+_21738194 | 0.96 |
ENSDART00000163688
|
FP085428.1
|
Danio rerio si:ch211-154o6.4 (si:ch211-154o6.4), mRNA. |
| chr16_-_54455573 | 0.96 |
ENSDART00000075275
|
pklr
|
pyruvate kinase L/R |
| chr7_-_24520866 | 0.95 |
ENSDART00000077039
|
faah2b
|
fatty acid amide hydrolase 2b |
| chr17_-_22048233 | 0.94 |
ENSDART00000155203
|
ttbk1b
|
tau tubulin kinase 1b |
| chr3_-_62527675 | 0.94 |
ENSDART00000155048
ENSDART00000064500 |
sox9b
|
SRY (sex determining region Y)-box 9b |
| chr7_+_29955368 | 0.93 |
ENSDART00000173686
|
tpma
|
alpha-tropomyosin |
| chr24_-_17047918 | 0.93 |
ENSDART00000020204
|
msrb2
|
methionine sulfoxide reductase B2 |
| chr7_+_30787903 | 0.93 |
ENSDART00000174000
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr15_+_6109861 | 0.92 |
ENSDART00000185154
|
PCP4 (1 of many)
|
Purkinje cell protein 4 |
| chr14_-_21219659 | 0.91 |
ENSDART00000089867
|
ppp2r2cb
|
protein phosphatase 2, regulatory subunit B, gamma b |
| chr23_-_7799184 | 0.90 |
ENSDART00000190946
ENSDART00000165427 |
myt1b
|
myelin transcription factor 1b |
| chr11_+_42730639 | 0.90 |
ENSDART00000165297
|
zgc:194981
|
zgc:194981 |
| chr14_+_25817246 | 0.89 |
ENSDART00000136733
|
glra1
|
glycine receptor, alpha 1 |
| chr3_+_26081343 | 0.89 |
ENSDART00000134647
|
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr19_+_1184878 | 0.88 |
ENSDART00000163539
|
scrt1a
|
scratch family zinc finger 1a |
| chr7_+_31879986 | 0.87 |
ENSDART00000138491
|
mybpc3
|
myosin binding protein C, cardiac |
| chr15_+_32711663 | 0.87 |
ENSDART00000157854
ENSDART00000167515 |
postnb
|
periostin, osteoblast specific factor b |
| chr25_+_31405266 | 0.86 |
ENSDART00000103395
|
tnnt3a
|
troponin T type 3a (skeletal, fast) |
| chr18_-_50862939 | 0.86 |
ENSDART00000180407
|
CABZ01113373.1
|
|
| chr1_+_17900306 | 0.86 |
ENSDART00000089480
|
cyp4v8
|
cytochrome P450, family 4, subfamily V, polypeptide 8 |
| chr2_-_9646857 | 0.86 |
ENSDART00000056901
|
zgc:153615
|
zgc:153615 |
| chr1_-_14332283 | 0.86 |
ENSDART00000090025
|
wfs1a
|
Wolfram syndrome 1a (wolframin) |
| chr9_+_42066030 | 0.85 |
ENSDART00000185311
ENSDART00000015267 |
pcbp3
|
poly(rC) binding protein 3 |
| chr21_-_27010796 | 0.85 |
ENSDART00000065398
ENSDART00000144342 ENSDART00000126542 |
ppp1r14ba
|
protein phosphatase 1, regulatory (inhibitor) subunit 14Ba |
| chr13_+_1542493 | 0.85 |
ENSDART00000181968
|
CABZ01044281.1
|
|
| chr4_-_1360495 | 0.84 |
ENSDART00000164623
|
ptn
|
pleiotrophin |
| chr21_+_19008168 | 0.83 |
ENSDART00000136196
ENSDART00000128381 ENSDART00000176624 |
nefla
|
neurofilament, light polypeptide a |
| chr19_-_21716593 | 0.81 |
ENSDART00000155126
|
znf516
|
zinc finger protein 516 |
| chr19_+_56351 | 0.81 |
ENSDART00000168334
|
col14a1b
|
collagen, type XIV, alpha 1b |
| chr2_+_37750016 | 0.81 |
ENSDART00000154726
|
si:dkeyp-66d1.7
|
si:dkeyp-66d1.7 |
| chr3_-_41791178 | 0.80 |
ENSDART00000049687
|
grifin
|
galectin-related inter-fiber protein |
| chr12_+_17865374 | 0.80 |
ENSDART00000169019
|
tmem130
|
transmembrane protein 130 |
| chr12_-_19103490 | 0.80 |
ENSDART00000060561
|
csdc2a
|
cold shock domain containing C2, RNA binding a |
| chr19_-_28789404 | 0.80 |
ENSDART00000191453
ENSDART00000026992 |
sox4a
|
SRY (sex determining region Y)-box 4a |
| chr8_-_21268303 | 0.79 |
ENSDART00000067211
|
gpr37l1b
|
G protein-coupled receptor 37 like 1b |
| chr21_-_10773344 | 0.78 |
ENSDART00000063244
|
grp
|
gastrin-releasing peptide |
| chr11_+_23957440 | 0.78 |
ENSDART00000190721
|
cntn2
|
contactin 2 |
| chr13_-_27675212 | 0.77 |
ENSDART00000141035
|
rims1a
|
regulating synaptic membrane exocytosis 1a |
| chr3_-_46818001 | 0.77 |
ENSDART00000166505
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr18_+_783936 | 0.77 |
ENSDART00000193357
|
rpp25b
|
ribonuclease P and MRP subunit p25, b |
| chr23_+_40460333 | 0.77 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr4_+_6643421 | 0.76 |
ENSDART00000099462
|
gpr85
|
G protein-coupled receptor 85 |
| chr1_-_21409877 | 0.76 |
ENSDART00000102782
|
gria2a
|
glutamate receptor, ionotropic, AMPA 2a |
| chr16_-_28856112 | 0.76 |
ENSDART00000078543
|
syt11b
|
synaptotagmin XIb |
| chr22_+_18389271 | 0.75 |
ENSDART00000088270
|
yjefn3
|
YjeF N-terminal domain containing 3 |
| chr16_-_26296477 | 0.74 |
ENSDART00000157553
|
erfl1
|
Ets2 repressor factor like 1 |
| chr6_-_13187168 | 0.73 |
ENSDART00000193286
ENSDART00000188350 ENSDART00000150036 ENSDART00000149940 |
adam23a
|
ADAM metallopeptidase domain 23a |
| chr10_-_32920690 | 0.71 |
ENSDART00000136245
|
cux1a
|
cut-like homeobox 1a |
| chr13_-_9318891 | 0.71 |
ENSDART00000137364
|
si:dkey-33c12.3
|
si:dkey-33c12.3 |
| chr5_+_55626693 | 0.70 |
ENSDART00000168908
ENSDART00000161412 |
ntrk2b
|
neurotrophic tyrosine kinase, receptor, type 2b |
| chr6_-_39764995 | 0.70 |
ENSDART00000085277
|
pfkmb
|
phosphofructokinase, muscle b |
| chr14_+_50770537 | 0.70 |
ENSDART00000158723
|
sncb
|
synuclein, beta |
| chr4_+_26628822 | 0.70 |
ENSDART00000191030
ENSDART00000186113 ENSDART00000186764 ENSDART00000165158 |
iqsec3a
|
IQ motif and Sec7 domain 3a |
| chr19_-_3759142 | 0.69 |
ENSDART00000170431
|
btr21
|
bloodthirsty-related gene family, member 21 |
| chr13_+_31769809 | 0.68 |
ENSDART00000147222
|
hif1aa
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) a |
| chr9_+_56422311 | 0.68 |
ENSDART00000171958
|
gpr39
|
G protein-coupled receptor 39 |
| chr9_+_34641237 | 0.68 |
ENSDART00000133996
|
shox
|
short stature homeobox |
| chr10_-_35542071 | 0.68 |
ENSDART00000162139
|
si:ch211-244c8.4
|
si:ch211-244c8.4 |
| chr25_-_169291 | 0.67 |
ENSDART00000128344
|
lipcb
|
lipase, hepatic b |
| chr1_-_49932040 | 0.67 |
ENSDART00000048281
|
cyp2u1
|
cytochrome P450, family 2, subfamily U, polypeptide 1 |
| chr20_+_22681066 | 0.67 |
ENSDART00000143286
|
lnx1
|
ligand of numb-protein X 1 |
| chr19_+_15571290 | 0.67 |
ENSDART00000131134
|
foxo6b
|
forkhead box O6 b |
| chr1_+_54137089 | 0.66 |
ENSDART00000062945
|
LO017798.1
|
|
| chr18_+_402048 | 0.66 |
ENSDART00000166345
|
gpib
|
glucose-6-phosphate isomerase b |
| chr5_-_37157510 | 0.66 |
ENSDART00000166710
|
lrch2
|
leucine-rich repeats and calponin homology (CH) domain containing 2 |
| chr13_+_4405282 | 0.66 |
ENSDART00000148280
|
prr18
|
proline rich 18 |
| chr5_+_38276582 | 0.66 |
ENSDART00000158532
|
gnb2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr9_-_18877597 | 0.65 |
ENSDART00000099446
|
kctd4
|
potassium channel tetramerization domain containing 4 |
| chr6_+_49723289 | 0.65 |
ENSDART00000190452
|
stx16
|
syntaxin 16 |
| chr18_+_40572294 | 0.65 |
ENSDART00000147003
|
si:ch211-132b12.2
|
si:ch211-132b12.2 |
| chr19_+_37925616 | 0.64 |
ENSDART00000148348
|
nxph1
|
neurexophilin 1 |
| chr11_+_3254252 | 0.64 |
ENSDART00000123568
|
pmela
|
premelanosome protein a |
| chr17_+_53425730 | 0.64 |
ENSDART00000127617
|
fabp10b
|
fatty acid binding protein 10b, liver basic |
| chr18_-_46208581 | 0.63 |
ENSDART00000141278
|
si:ch211-14c7.2
|
si:ch211-14c7.2 |
| chr2_-_5074812 | 0.63 |
ENSDART00000163728
|
dlg1l
|
discs, large (Drosophila) homolog 1, like |
| chr4_+_16323970 | 0.63 |
ENSDART00000190651
|
BX322608.1
|
|
| chr12_+_45676667 | 0.62 |
ENSDART00000016553
|
si:ch73-111m19.2
|
si:ch73-111m19.2 |
| chr20_-_54462551 | 0.62 |
ENSDART00000171769
ENSDART00000169692 |
evlb
|
Enah/Vasp-like b |
| chr5_-_36837846 | 0.62 |
ENSDART00000032481
|
ckma
|
creatine kinase, muscle a |
| chr10_+_158590 | 0.62 |
ENSDART00000081982
|
KCNJ15
|
potassium voltage-gated channel subfamily J member 15 |
| chr11_-_141592 | 0.62 |
ENSDART00000092787
|
cdk4
|
cyclin-dependent kinase 4 |
| chr13_-_29424454 | 0.62 |
ENSDART00000026765
|
slc18a3a
|
solute carrier family 18 (vesicular acetylcholine transporter), member 3a |
| chr5_-_22501663 | 0.62 |
ENSDART00000133174
|
si:dkey-27p18.5
|
si:dkey-27p18.5 |
| chr7_-_49594995 | 0.61 |
ENSDART00000174161
ENSDART00000109147 |
brsk2b
|
BR serine/threonine kinase 2b |
| chr10_-_5847904 | 0.61 |
ENSDART00000161096
|
ankrd55
|
ankyrin repeat domain 55 |
| chr20_-_26001288 | 0.61 |
ENSDART00000136518
ENSDART00000063177 |
capn3b
|
calpain 3b |
| chr17_-_15528597 | 0.61 |
ENSDART00000150232
|
fyna
|
FYN proto-oncogene, Src family tyrosine kinase a |
| chr3_-_46817838 | 0.61 |
ENSDART00000028610
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr20_-_34868814 | 0.61 |
ENSDART00000153049
|
stmn4
|
stathmin-like 4 |
| chr2_-_985417 | 0.61 |
ENSDART00000140540
|
si:ch211-241e1.3
|
si:ch211-241e1.3 |
| chr9_+_13707933 | 0.60 |
ENSDART00000141194
|
mpp4a
|
membrane protein, palmitoylated 4a (MAGUK p55 subfamily member 4) |
| chr12_-_4388704 | 0.60 |
ENSDART00000152168
|
si:ch211-173d10.1
|
si:ch211-173d10.1 |
| chr14_+_22172047 | 0.60 |
ENSDART00000114750
ENSDART00000148259 |
gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr11_-_37509001 | 0.60 |
ENSDART00000109753
|
bsnb
|
bassoon (presynaptic cytomatrix protein) b |
| chr21_+_21374277 | 0.59 |
ENSDART00000079431
|
rtn2b
|
reticulon 2b |
| chr22_-_354592 | 0.59 |
ENSDART00000155769
|
tmem240b
|
transmembrane protein 240b |
| chr7_-_67894046 | 0.59 |
ENSDART00000168964
|
si:ch73-315f9.2
|
si:ch73-315f9.2 |
| chr14_+_21783229 | 0.58 |
ENSDART00000170784
|
ankrd13d
|
ankyrin repeat domain 13 family, member D |
| chr5_-_68916455 | 0.58 |
ENSDART00000171465
|
ank1a
|
ankyrin 1, erythrocytic a |
| chr3_+_59051503 | 0.58 |
ENSDART00000160767
|
rasd4
|
rasd family member 4 |
| chr14_-_1454045 | 0.58 |
ENSDART00000161460
|
pmt
|
phosphoethanolamine methyltransferase |
| chr13_+_24552254 | 0.58 |
ENSDART00000147907
|
lgalslb
|
lectin, galactoside-binding-like b |
| chr3_-_57744323 | 0.58 |
ENSDART00000101829
|
lgals3bpb
|
lectin, galactoside-binding, soluble, 3 binding protein b |
| chr23_+_45579497 | 0.57 |
ENSDART00000110381
|
egr4
|
early growth response 4 |
| chr19_-_35229336 | 0.57 |
ENSDART00000054274
|
macf1a
|
microtubule-actin crosslinking factor 1a |
| chr14_+_8275115 | 0.57 |
ENSDART00000129055
|
nrg2b
|
neuregulin 2b |
| chr10_-_17103651 | 0.57 |
ENSDART00000108959
|
RNF208
|
ring finger protein 208 |
| chr19_-_6239248 | 0.56 |
ENSDART00000014127
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr11_+_3254524 | 0.56 |
ENSDART00000159459
|
pmela
|
premelanosome protein a |
| chr23_-_24582606 | 0.56 |
ENSDART00000129910
|
tmem240a
|
transmembrane protein 240a |
| chr16_-_24518027 | 0.55 |
ENSDART00000134120
ENSDART00000143761 |
cadm4
|
cell adhesion molecule 4 |
| chr5_-_55395384 | 0.55 |
ENSDART00000147298
ENSDART00000082577 |
prune2
|
prune homolog 2 (Drosophila) |
| chr7_-_56793739 | 0.55 |
ENSDART00000082842
|
si:ch211-146m13.3
|
si:ch211-146m13.3 |
| chr19_-_9472893 | 0.55 |
ENSDART00000045565
ENSDART00000137505 |
vamp1
|
vesicle-associated membrane protein 1 |
| chr4_+_21129752 | 0.55 |
ENSDART00000169764
|
syt1a
|
synaptotagmin Ia |
| chr6_-_138392 | 0.55 |
ENSDART00000148974
|
keap1b
|
kelch-like ECH-associated protein 1b |
| chr2_+_30916188 | 0.55 |
ENSDART00000137012
|
myom1a
|
myomesin 1a (skelemin) |
| chr1_-_43712120 | 0.55 |
ENSDART00000074600
|
SLC9B2
|
si:dkey-162b23.4 |
| chr11_+_11201096 | 0.53 |
ENSDART00000171916
ENSDART00000171521 ENSDART00000087105 ENSDART00000159603 |
myom2a
|
myomesin 2a |
| chr11_-_44873780 | 0.53 |
ENSDART00000160465
|
opn6a
|
opsin 6, group member a |
| chr12_-_41684729 | 0.53 |
ENSDART00000184461
|
jakmip3
|
Janus kinase and microtubule interacting protein 3 |
| chr20_-_14680897 | 0.53 |
ENSDART00000063857
ENSDART00000161314 |
scrn2
|
secernin 2 |
| chr14_-_2285955 | 0.52 |
ENSDART00000183928
|
pcdh2ab9
|
protocadherin 2 alpha b 9 |
| chr12_+_23875318 | 0.52 |
ENSDART00000152869
|
svila
|
supervillin a |
| chr1_-_59417949 | 0.52 |
ENSDART00000170558
|
si:ch211-188p14.3
|
si:ch211-188p14.3 |
| chr11_+_40649412 | 0.52 |
ENSDART00000043016
ENSDART00000134560 |
slc45a1
|
solute carrier family 45, member 1 |
| chr3_+_31396149 | 0.52 |
ENSDART00000151423
ENSDART00000193580 |
c1ql3b
|
complement component 1, q subcomponent-like 3b |
| chr14_-_39031108 | 0.51 |
ENSDART00000026194
|
glra4a
|
glycine receptor, alpha 4a |
| chr8_+_694218 | 0.51 |
ENSDART00000147753
|
rnf165b
|
ring finger protein 165b |
| chr13_+_31180084 | 0.51 |
ENSDART00000133774
|
ptpn20
|
protein tyrosine phosphatase, non-receptor type 20 |
| chr5_+_72108241 | 0.51 |
ENSDART00000006606
|
fabp1a
|
fatty acid binding protein 1a, liver |
| chr19_-_9662958 | 0.51 |
ENSDART00000041094
|
clcn1a
|
chloride channel, voltage-sensitive 1a |
| chr3_+_58231560 | 0.51 |
ENSDART00000126505
|
CABZ01038494.1
|
|
| chr6_-_138603 | 0.51 |
ENSDART00000148911
|
keap1b
|
kelch-like ECH-associated protein 1b |
| chr20_+_54383838 | 0.51 |
ENSDART00000157737
|
lrfn5b
|
leucine rich repeat and fibronectin type III domain containing 5b |
| chr17_+_53250802 | 0.51 |
ENSDART00000143819
|
VASH1
|
vasohibin 1 |
| chr17_-_50331351 | 0.51 |
ENSDART00000149294
|
otofb
|
otoferlin b |
| chr1_-_22687913 | 0.51 |
ENSDART00000168171
|
fgfbp2b
|
fibroblast growth factor binding protein 2b |
| chr20_-_31075972 | 0.50 |
ENSDART00000122927
|
si:ch211-198b3.4
|
si:ch211-198b3.4 |
| chr13_-_226109 | 0.50 |
ENSDART00000161705
ENSDART00000172744 ENSDART00000163902 ENSDART00000158208 |
rtn4b
|
reticulon 4b |
| chr16_-_43025885 | 0.50 |
ENSDART00000193146
ENSDART00000157302 |
si:dkey-7j14.5
|
si:dkey-7j14.5 |
| chr10_+_21730585 | 0.50 |
ENSDART00000188576
|
pcdh1g22
|
protocadherin 1 gamma 22 |
| chr14_+_25816874 | 0.50 |
ENSDART00000005499
|
glra1
|
glycine receptor, alpha 1 |
| chr3_-_55147731 | 0.50 |
ENSDART00000155871
ENSDART00000109016 ENSDART00000122904 |
hbae3
|
hemoglobin alpha embryonic-3 |
| chr13_-_37180815 | 0.50 |
ENSDART00000139907
|
si:dkeyp-77c8.1
|
si:dkeyp-77c8.1 |
| chr13_+_27951688 | 0.49 |
ENSDART00000050303
|
b3gat2
|
beta-1,3-glucuronyltransferase 2 (glucuronosyltransferase S) |
| chr5_-_19394440 | 0.49 |
ENSDART00000163771
|
foxn4
|
forkhead box N4 |
| chr6_+_27452289 | 0.49 |
ENSDART00000186265
|
sned1
|
sushi, nidogen and EGF-like domains 1 |
| chr10_-_15128771 | 0.49 |
ENSDART00000101261
|
spp1
|
secreted phosphoprotein 1 |
| chr23_+_4299887 | 0.49 |
ENSDART00000132604
|
l3mbtl1a
|
l(3)mbt-like 1a (Drosophila) |
| chr8_+_23165749 | 0.48 |
ENSDART00000063057
|
dnajc5aa
|
DnaJ (Hsp40) homolog, subfamily C, member 5aa |
| chr13_+_255067 | 0.48 |
ENSDART00000102505
|
foxg1d
|
forkhead box G1d |
| chr5_+_21931124 | 0.48 |
ENSDART00000137627
|
si:ch73-92i20.1
|
si:ch73-92i20.1 |
| chr11_-_103136 | 0.48 |
ENSDART00000173308
ENSDART00000162982 |
elmo2
|
engulfment and cell motility 2 |
| chr8_+_28065803 | 0.48 |
ENSDART00000178481
|
kcnd3
|
potassium voltage-gated channel, Shal-related subfamily, member 3 |
| chr10_-_5847655 | 0.48 |
ENSDART00000192773
|
ankrd55
|
ankyrin repeat domain 55 |
| chr23_+_37323962 | 0.48 |
ENSDART00000102881
|
fam43b
|
family with sequence similarity 43, member B |
| chr5_-_55395964 | 0.47 |
ENSDART00000145791
|
prune2
|
prune homolog 2 (Drosophila) |
| chr6_+_27146671 | 0.47 |
ENSDART00000156792
|
kif1aa
|
kinesin family member 1Aa |
Network of associatons between targets according to the STRING database.
First level regulatory network of pax2a+pax5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.5 | 1.4 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.4 | 1.2 | GO:0097435 | fibril organization(GO:0097435) |
| 0.4 | 1.5 | GO:0090387 | phagosome maturation involved in apoptotic cell clearance(GO:0090386) phagolysosome assembly involved in apoptotic cell clearance(GO:0090387) |
| 0.3 | 1.0 | GO:1903792 | regulation of neurotransmitter uptake(GO:0051580) negative regulation of anion transport(GO:1903792) |
| 0.3 | 0.9 | GO:0071435 | potassium ion export(GO:0071435) potassium ion export across plasma membrane(GO:0097623) |
| 0.3 | 2.9 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.3 | 0.9 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.3 | 0.8 | GO:0048677 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.3 | 1.0 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.2 | 0.7 | GO:1990416 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.2 | 0.9 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.2 | 0.9 | GO:0030091 | protein repair(GO:0030091) |
| 0.2 | 2.3 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.2 | 0.9 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.2 | 0.9 | GO:0031446 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 0.2 | 1.3 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.2 | 0.6 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.2 | 0.6 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.2 | 1.2 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.2 | 0.7 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.2 | 2.0 | GO:0061615 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.2 | 0.9 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.2 | 0.8 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.2 | 0.5 | GO:0021961 | posterior commissure morphogenesis(GO:0021961) |
| 0.2 | 0.8 | GO:0034653 | vitamin catabolic process(GO:0009111) diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) fat-soluble vitamin catabolic process(GO:0042363) |
| 0.1 | 0.9 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.1 | 0.6 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.1 | 0.4 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 0.4 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.1 | 0.8 | GO:0086091 | regulation of heart rate by cardiac conduction(GO:0086091) |
| 0.1 | 0.5 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 0.1 | 0.7 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.1 | 0.3 | GO:0048917 | posterior lateral line ganglion development(GO:0048917) |
| 0.1 | 1.1 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.1 | 0.6 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.1 | 0.4 | GO:1903385 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 1.0 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) |
| 0.1 | 0.5 | GO:0032655 | interleukin-12 production(GO:0032615) regulation of interleukin-12 production(GO:0032655) |
| 0.1 | 1.0 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.1 | 0.2 | GO:0035992 | tendon formation(GO:0035992) |
| 0.1 | 0.4 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.1 | 0.3 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.1 | 0.2 | GO:0014856 | skeletal muscle cell proliferation(GO:0014856) regulation of skeletal muscle cell proliferation(GO:0014857) |
| 0.1 | 0.5 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.1 | 0.2 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 0.6 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 0.1 | GO:0090183 | regulation of morphogenesis of a branching structure(GO:0060688) positive regulation of mesonephros development(GO:0061213) regulation of mesonephros development(GO:0061217) regulation of kidney development(GO:0090183) positive regulation of kidney development(GO:0090184) regulation of branching involved in ureteric bud morphogenesis(GO:0090189) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.1 | 0.2 | GO:0036336 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.1 | 0.3 | GO:0055014 | atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 0.3 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.1 | 1.2 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.1 | 0.3 | GO:0071675 | regulation of macrophage chemotaxis(GO:0010758) positive regulation of macrophage chemotaxis(GO:0010759) regulation of odontogenesis(GO:0042481) mononuclear cell migration(GO:0071674) regulation of mononuclear cell migration(GO:0071675) |
| 0.1 | 0.4 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 0.2 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 0.7 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 0.8 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 0.2 | GO:0045922 | negative regulation of fatty acid metabolic process(GO:0045922) |
| 0.1 | 0.6 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.1 | 0.5 | GO:0072098 | anterior/posterior pattern specification involved in pronephros development(GO:0034672) anterior/posterior pattern specification involved in kidney development(GO:0072098) |
| 0.1 | 0.8 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.1 | 0.3 | GO:0032689 | negative regulation of interferon-gamma production(GO:0032689) CD4-positive, alpha-beta T cell proliferation(GO:0035739) regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.1 | 0.2 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.1 | 1.2 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.3 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.6 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 6.9 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.1 | 0.3 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.0 | 0.8 | GO:0039014 | pronephric nephron tubule epithelial cell differentiation(GO:0035778) cell differentiation involved in pronephros development(GO:0039014) nephron tubule epithelial cell differentiation(GO:0072160) |
| 0.0 | 0.3 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.0 | 0.2 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.4 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.6 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 1.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.6 | GO:0006599 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 0.8 | GO:0048899 | anterior lateral line development(GO:0048899) |
| 0.0 | 0.6 | GO:0001964 | startle response(GO:0001964) |
| 0.0 | 0.3 | GO:0003222 | ventricular trabecula myocardium morphogenesis(GO:0003222) |
| 0.0 | 0.3 | GO:0021627 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.0 | 0.1 | GO:0043903 | regulation of symbiosis, encompassing mutualism through parasitism(GO:0043903) |
| 0.0 | 0.4 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.0 | 0.4 | GO:0071322 | cellular response to carbohydrate stimulus(GO:0071322) cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.0 | 0.1 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.4 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.2 | GO:0098743 | cartilage condensation(GO:0001502) cell aggregation(GO:0098743) |
| 0.0 | 0.3 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.0 | 0.2 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 1.0 | GO:0007257 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 4.4 | GO:0061515 | myeloid cell development(GO:0061515) |
| 0.0 | 0.9 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.2 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.0 | 0.5 | GO:0060035 | notochord cell development(GO:0060035) |
| 0.0 | 0.3 | GO:0032196 | transposition(GO:0032196) |
| 0.0 | 0.9 | GO:0006942 | regulation of striated muscle contraction(GO:0006942) |
| 0.0 | 0.4 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.7 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.0 | 1.1 | GO:0002068 | glandular epithelial cell development(GO:0002068) |
| 0.0 | 0.3 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.0 | 0.2 | GO:0090134 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.0 | 0.1 | GO:1901004 | ubiquinone-6 metabolic process(GO:1901004) ubiquinone-6 biosynthetic process(GO:1901006) |
| 0.0 | 0.2 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.7 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.3 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.3 | GO:2000463 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.0 | 0.2 | GO:0045190 | B cell activation involved in immune response(GO:0002312) isotype switching(GO:0045190) |
| 0.0 | 0.2 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.2 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.0 | 0.8 | GO:1902807 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.3 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.1 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.6 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.8 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.0 | 1.3 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.4 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.2 | GO:0010460 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) |
| 0.0 | 3.0 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 1.5 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.4 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.0 | 0.1 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.0 | 0.9 | GO:0048885 | neuromast deposition(GO:0048885) |
| 0.0 | 0.3 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.4 | GO:0021554 | optic nerve development(GO:0021554) |
| 0.0 | 0.3 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.7 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.0 | 0.3 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 0.0 | 0.1 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.0 | 0.7 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.4 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.0 | 2.1 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.4 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.0 | 0.3 | GO:1990118 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.4 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.0 | 0.1 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 | 0.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 1.1 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.4 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.2 | GO:0097241 | hematopoietic stem cell migration to bone marrow(GO:0097241) |
| 0.0 | 0.4 | GO:0000272 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 1.3 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.3 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.4 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.7 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.8 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.3 | GO:0019883 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.8 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:1901881 | positive regulation of protein depolymerization(GO:1901881) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.5 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.3 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.4 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.2 | GO:0019430 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.4 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.1 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.0 | 0.5 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 0.1 | GO:0019557 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.1 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.0 | 0.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.3 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.2 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.0 | GO:0043363 | nucleate erythrocyte differentiation(GO:0043363) |
| 0.0 | 1.1 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 0.2 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.2 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.4 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.3 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.1 | GO:0090075 | relaxation of muscle(GO:0090075) |
| 0.0 | 0.3 | GO:0003417 | endochondral bone growth(GO:0003416) growth plate cartilage development(GO:0003417) bone growth(GO:0098868) |
| 0.0 | 1.2 | GO:0030837 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.0 | 0.6 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.2 | GO:0060034 | notochord cell differentiation(GO:0060034) |
| 0.0 | 0.2 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.4 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.2 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.0 | 0.4 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.3 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.8 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.8 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.3 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.1 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.1 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.3 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.0 | 3.4 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 0.2 | GO:0006337 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.1 | GO:0071632 | optomotor response(GO:0071632) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.0 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.0 | 0.6 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.3 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.2 | GO:0006032 | chitin catabolic process(GO:0006032) glucosamine-containing compound catabolic process(GO:1901072) |
| 0.0 | 0.0 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.1 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.7 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 1.0 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.1 | GO:0042779 | tRNA 3'-trailer cleavage(GO:0042779) |
| 0.0 | 0.2 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.0 | 0.4 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 1.5 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.2 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.2 | GO:0099638 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.0 | 0.0 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.0 | 2.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.1 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.0 | 0.7 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.1 | GO:0070166 | tooth mineralization(GO:0034505) enamel mineralization(GO:0070166) |
| 0.0 | 0.1 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.2 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.1 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.1 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.0 | 0.9 | GO:0033339 | pectoral fin development(GO:0033339) |
| 0.0 | 0.0 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 0.3 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.0 | GO:0044783 | G1 DNA damage checkpoint(GO:0044783) |
| 0.0 | 0.3 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.2 | GO:0038034 | signal transduction in absence of ligand(GO:0038034) extrinsic apoptotic signaling pathway in absence of ligand(GO:0097192) |
| 0.0 | 0.3 | GO:0046514 | ceramide catabolic process(GO:0046514) |
| 0.0 | 0.3 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.0 | 0.2 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
| 0.0 | 0.5 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.1 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 1.1 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.5 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.1 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.0 | 0.1 | GO:0035188 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.1 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.1 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.0 | 1.2 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 0.5 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 0.2 | GO:0033338 | medial fin development(GO:0033338) |
| 0.0 | 0.1 | GO:0001574 | ganglioside metabolic process(GO:0001573) ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.2 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.1 | GO:0070509 | calcium ion import(GO:0070509) |
| 0.0 | 0.1 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.0 | 0.5 | GO:0060348 | bone development(GO:0060348) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.3 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.2 | 0.9 | GO:0031673 | H zone(GO:0031673) |
| 0.2 | 1.3 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.2 | 2.0 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.2 | 1.1 | GO:0016234 | inclusion body(GO:0016234) |
| 0.1 | 2.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 1.7 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.4 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.1 | 0.8 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 0.4 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 1.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.4 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.1 | 0.5 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 0.6 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 0.6 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.1 | 2.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 1.4 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 0.3 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.1 | 0.3 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.3 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 1.0 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.8 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.0 | 4.8 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.7 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 1.4 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 3.6 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 0.3 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.2 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 3.0 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.9 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.2 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.0 | 1.1 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.5 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.6 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.5 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.7 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 1.5 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 1.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.4 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.1 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 0.4 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 1.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.3 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 1.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.7 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 2.8 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.0 | GO:0008352 | katanin complex(GO:0008352) |
| 0.0 | 1.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.1 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.1 | GO:0032426 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) stereocilium tip(GO:0032426) |
| 0.0 | 0.4 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.4 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 4.2 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.2 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.2 | 0.7 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.2 | 0.9 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.2 | 0.7 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.2 | 1.1 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.2 | 0.7 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.2 | 1.3 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.2 | 3.6 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 1.7 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.2 | 2.0 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.2 | 1.0 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 1.0 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.1 | 1.3 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 1.1 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 1.0 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 2.3 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.1 | 0.5 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.1 | 1.7 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 0.3 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.1 | 0.5 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.1 | 0.3 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 0.8 | GO:0005351 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 1.0 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.4 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 0.4 | GO:0004135 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.1 | 0.8 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.9 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 1.0 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.1 | 0.3 | GO:0015105 | arsenite transmembrane transporter activity(GO:0015105) |
| 0.1 | 0.8 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 6.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.4 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 0.3 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 0.3 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 0.4 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 0.8 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 0.8 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.1 | 0.6 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.4 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.1 | 0.2 | GO:0047173 | phosphatidylcholine-retinol O-acyltransferase activity(GO:0047173) |
| 0.1 | 0.6 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.3 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.6 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.1 | 0.2 | GO:0010852 | cyclase inhibitor activity(GO:0010852) guanylate cyclase inhibitor activity(GO:0030251) |
| 0.1 | 0.8 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.6 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.1 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.2 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.0 | 0.6 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.6 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 0.4 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.6 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.2 | GO:0048531 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.2 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.9 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.3 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 1.1 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.4 | GO:0015386 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.6 | GO:0008443 | 6-phosphofructo-2-kinase activity(GO:0003873) phosphofructokinase activity(GO:0008443) |
| 0.0 | 0.3 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.9 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.4 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.0 | 1.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.2 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 1.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.3 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.0 | 0.2 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.5 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.5 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 1.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.1 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.0 | 0.3 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.1 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.4 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.5 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.4 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.2 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.4 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.0 | 0.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 1.7 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.8 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.3 | GO:0022821 | calcium, potassium:sodium antiporter activity(GO:0008273) potassium ion antiporter activity(GO:0022821) |
| 0.0 | 0.2 | GO:0004960 | thromboxane receptor activity(GO:0004960) |
| 0.0 | 0.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.9 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 4.1 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.1 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.0 | 0.1 | GO:1902945 | metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902945) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.6 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.3 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0031704 | apelin receptor binding(GO:0031704) |
| 0.0 | 0.5 | GO:0008308 | voltage-gated chloride channel activity(GO:0005247) voltage-gated anion channel activity(GO:0008308) |
| 0.0 | 1.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.2 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.3 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.4 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 1.0 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.2 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.9 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.3 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.8 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.5 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.1 | GO:0102344 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.0 | 0.1 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.0 | 0.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.3 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.3 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.3 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.2 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.1 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.5 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.2 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.1 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.9 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.3 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 1.7 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.1 | GO:2001070 | starch binding(GO:2001070) |
| 0.0 | 0.0 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.0 | 0.0 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.0 | 1.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.5 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.3 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.4 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.5 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.0 | 0.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.4 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.1 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.4 | GO:1990939 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) ATP-dependent microtubule motor activity(GO:1990939) |
| 0.0 | 0.1 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.3 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.3 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 0.1 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.7 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.2 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.4 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.4 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.4 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.1 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.6 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.4 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.3 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.4 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.2 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.4 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.2 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.4 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.1 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.3 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.3 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.2 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.6 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 1.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 1.5 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 2.2 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 0.8 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 1.0 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 0.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 1.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.1 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 1.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 0.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 0.5 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.9 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.3 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 1.0 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.5 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.4 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.4 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.4 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.5 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.9 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.0 | 0.9 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.6 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.5 | REACTOME ION CHANNEL TRANSPORT | Genes involved in Ion channel transport |
| 0.0 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.3 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |