Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
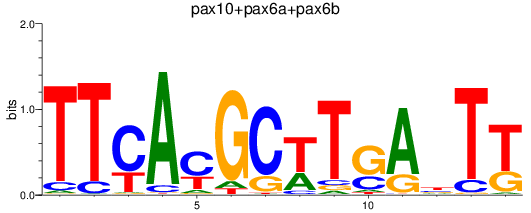
Results for pax10+pax6a+pax6b
Z-value: 0.63
Transcription factors associated with pax10+pax6a+pax6b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pax6b
|
ENSDARG00000045936 | paired box 6b |
|
pax10
|
ENSDARG00000053364 | paired box 10 |
|
pax6a
|
ENSDARG00000103379 | paired box 6a |
|
pax10
|
ENSDARG00000111379 | paired box 10 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pax10 | dr11_v1_chr3_+_32553714_32553714 | 0.59 | 7.7e-03 | Click! |
| pax6a | dr11_v1_chr25_-_15040369_15040442 | 0.56 | 1.2e-02 | Click! |
| pax6b | dr11_v1_chr7_+_15871156_15871156 | 0.21 | 3.9e-01 | Click! |
Activity profile of pax10+pax6a+pax6b motif
Sorted Z-values of pax10+pax6a+pax6b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_8053385 | 1.77 |
ENSDART00000142714
|
si:ch211-251f6.7
|
si:ch211-251f6.7 |
| chr10_-_8046764 | 1.51 |
ENSDART00000099031
|
zgc:136254
|
zgc:136254 |
| chr12_-_35949936 | 1.19 |
ENSDART00000192583
|
AL954682.1
|
|
| chr10_-_8053753 | 1.19 |
ENSDART00000162289
|
si:ch211-251f6.7
|
si:ch211-251f6.7 |
| chr25_-_27842654 | 1.03 |
ENSDART00000154852
ENSDART00000156906 |
asb15a
|
ankyrin repeat and SOCS box containing 15a |
| chr13_-_22862133 | 0.89 |
ENSDART00000138563
|
pbld2
|
phenazine biosynthesis-like protein domain containing 2 |
| chr2_-_32768951 | 0.78 |
ENSDART00000004712
|
bfsp2
|
beaded filament structural protein 2, phakinin |
| chr16_-_31188715 | 0.77 |
ENSDART00000058829
|
scrt1b
|
scratch family zinc finger 1b |
| chr13_-_32726178 | 0.72 |
ENSDART00000012232
|
pdss2
|
prenyl (decaprenyl) diphosphate synthase, subunit 2 |
| chr5_-_51619742 | 0.71 |
ENSDART00000188537
|
otpb
|
orthopedia homeobox b |
| chr22_+_18389271 | 0.67 |
ENSDART00000088270
|
yjefn3
|
YjeF N-terminal domain containing 3 |
| chr3_+_35298078 | 0.64 |
ENSDART00000110126
|
cacng3b
|
calcium channel, voltage-dependent, gamma subunit 3b |
| chr11_+_42730639 | 0.62 |
ENSDART00000165297
|
zgc:194981
|
zgc:194981 |
| chr21_+_23108420 | 0.60 |
ENSDART00000192394
ENSDART00000088459 |
htr3b
|
5-hydroxytryptamine (serotonin) receptor 3B |
| chr24_-_41320037 | 0.55 |
ENSDART00000129058
|
rheb
|
Ras homolog, mTORC1 binding |
| chr24_+_38306010 | 0.53 |
ENSDART00000143184
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr14_-_1454045 | 0.52 |
ENSDART00000161460
|
pmt
|
phosphoethanolamine methyltransferase |
| chr10_-_44306399 | 0.51 |
ENSDART00000180042
|
CDK2AP1
|
cyclin dependent kinase 2 associated protein 1 |
| chr5_+_32162684 | 0.50 |
ENSDART00000134472
|
taok3b
|
TAO kinase 3b |
| chr19_-_47526737 | 0.49 |
ENSDART00000186636
|
scg5
|
secretogranin V |
| chr9_+_31222026 | 0.49 |
ENSDART00000145573
|
clybl
|
citrate lyase beta like |
| chr5_+_23242370 | 0.48 |
ENSDART00000051532
|
agtr2
|
angiotensin II receptor, type 2 |
| chr20_-_29474859 | 0.48 |
ENSDART00000152906
ENSDART00000045249 |
scg5
|
secretogranin V |
| chr10_-_31015535 | 0.47 |
ENSDART00000146116
|
panx3
|
pannexin 3 |
| chr24_-_6671549 | 0.46 |
ENSDART00000114700
|
unm_sa821
|
un-named sa821 |
| chr25_+_19105804 | 0.46 |
ENSDART00000104414
|
rlbp1b
|
retinaldehyde binding protein 1b |
| chr1_-_12278056 | 0.45 |
ENSDART00000139336
ENSDART00000137463 |
cplx2l
|
complexin 2, like |
| chr7_+_21887787 | 0.45 |
ENSDART00000162252
|
pop7
|
POP7 homolog, ribonuclease P/MRP subunit |
| chr1_-_59287410 | 0.45 |
ENSDART00000158011
ENSDART00000170580 |
col5a3b
|
collagen, type V, alpha 3b |
| chr14_+_20893065 | 0.44 |
ENSDART00000079452
|
lygl1
|
lysozyme g-like 1 |
| chr12_-_44072716 | 0.44 |
ENSDART00000162509
|
si:ch73-329n5.3
|
si:ch73-329n5.3 |
| chr21_-_7178348 | 0.44 |
ENSDART00000187467
|
fam69b
|
family with sequence similarity 69, member B |
| chr7_+_26762958 | 0.44 |
ENSDART00000167956
ENSDART00000134717 |
tspan18a
|
tetraspanin 18a |
| chr18_-_20466061 | 0.44 |
ENSDART00000060311
|
paqr5a
|
progestin and adipoQ receptor family member Va |
| chr7_+_17229980 | 0.43 |
ENSDART00000184910
|
slc6a5
|
solute carrier family 6 (neurotransmitter transporter), member 5 |
| chr23_-_27505825 | 0.43 |
ENSDART00000137229
ENSDART00000013797 |
asb8
|
ankyrin repeat and SOCS box containing 8 |
| chr23_+_44611864 | 0.42 |
ENSDART00000145905
ENSDART00000132361 |
eno3
|
enolase 3, (beta, muscle) |
| chr25_+_19106574 | 0.42 |
ENSDART00000067332
|
rlbp1b
|
retinaldehyde binding protein 1b |
| chr19_-_41518922 | 0.42 |
ENSDART00000164483
ENSDART00000062080 |
chrac1
|
chromatin accessibility complex 1 |
| chr23_+_44614056 | 0.42 |
ENSDART00000188379
|
eno3
|
enolase 3, (beta, muscle) |
| chr10_+_26972755 | 0.42 |
ENSDART00000042162
|
tm7sf2
|
transmembrane 7 superfamily member 2 |
| chr19_-_9829965 | 0.41 |
ENSDART00000136842
ENSDART00000142766 |
cacng8a
|
calcium channel, voltage-dependent, gamma subunit 8a |
| chr3_-_26184018 | 0.40 |
ENSDART00000191604
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr2_+_6991208 | 0.40 |
ENSDART00000182239
|
ddr2b
|
discoidin domain receptor tyrosine kinase 2b |
| chr22_+_27090136 | 0.39 |
ENSDART00000136770
|
si:dkey-246e1.3
|
si:dkey-246e1.3 |
| chr19_+_12444943 | 0.39 |
ENSDART00000135706
|
ldlrad4a
|
low density lipoprotein receptor class A domain containing 4a |
| chr22_-_25033105 | 0.39 |
ENSDART00000124220
|
nptxrb
|
neuronal pentraxin receptor b |
| chr5_-_33039670 | 0.38 |
ENSDART00000141361
|
glipr2
|
GLI pathogenesis-related 2 |
| chr11_-_11336986 | 0.38 |
ENSDART00000016677
|
zgc:77929
|
zgc:77929 |
| chr6_+_11990733 | 0.38 |
ENSDART00000151075
|
baz2ba
|
bromodomain adjacent to zinc finger domain, 2Ba |
| chr8_+_26083808 | 0.38 |
ENSDART00000099283
|
dalrd3
|
DALR anticodon binding domain containing 3 |
| chr1_+_54908895 | 0.37 |
ENSDART00000145652
|
golga7ba
|
golgin A7 family, member Ba |
| chr6_+_42819337 | 0.37 |
ENSDART00000046498
|
sema3fa
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Fa |
| chr5_-_29750377 | 0.36 |
ENSDART00000051474
|
barhl1a
|
BarH-like homeobox 1a |
| chr3_+_43086548 | 0.36 |
ENSDART00000163579
|
si:dkey-43p13.5
|
si:dkey-43p13.5 |
| chr25_-_28926631 | 0.36 |
ENSDART00000112850
|
etfbkmt
|
electron transfer flavoprotein beta subunit lysine methyltransferase |
| chr20_-_14680897 | 0.36 |
ENSDART00000063857
ENSDART00000161314 |
scrn2
|
secernin 2 |
| chr20_+_36233873 | 0.35 |
ENSDART00000131867
|
cnih3
|
cornichon family AMPA receptor auxiliary protein 3 |
| chr14_+_3495542 | 0.35 |
ENSDART00000168934
|
gstp2
|
glutathione S-transferase pi 2 |
| chr23_-_36724575 | 0.35 |
ENSDART00000159560
|
agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr5_-_51756210 | 0.35 |
ENSDART00000163464
|
lhfpl2b
|
LHFPL tetraspan subfamily member 2b |
| chr2_-_985417 | 0.35 |
ENSDART00000140540
|
si:ch211-241e1.3
|
si:ch211-241e1.3 |
| chr11_-_6265574 | 0.35 |
ENSDART00000181974
ENSDART00000104405 |
ccl25b
|
chemokine (C-C motif) ligand 25b |
| chr9_+_23900703 | 0.35 |
ENSDART00000127859
|
trim63b
|
tripartite motif containing 63b |
| chr3_+_62353650 | 0.34 |
ENSDART00000112428
|
iqck
|
IQ motif containing K |
| chr11_+_24298432 | 0.34 |
ENSDART00000138487
|
si:dkey-76p14.2
|
si:dkey-76p14.2 |
| chr15_+_5901970 | 0.34 |
ENSDART00000114134
|
wrb
|
tryptophan rich basic protein |
| chr21_+_27382893 | 0.34 |
ENSDART00000005682
|
actn3a
|
actinin alpha 3a |
| chr12_-_10441661 | 0.34 |
ENSDART00000014379
|
uqcrc2b
|
ubiquinol-cytochrome c reductase core protein 2b |
| chr12_+_3571770 | 0.33 |
ENSDART00000164707
ENSDART00000189819 |
coa3a
|
cytochrome C oxidase assembly factor 3a |
| chr7_+_48555400 | 0.33 |
ENSDART00000174474
|
kcnq1
|
potassium voltage-gated channel, KQT-like subfamily, member 1 |
| chr14_-_14659023 | 0.33 |
ENSDART00000170355
ENSDART00000159888 ENSDART00000172241 |
nsdhl
|
NAD(P) dependent steroid dehydrogenase-like |
| chr24_-_8729531 | 0.33 |
ENSDART00000082346
|
tfap2a
|
transcription factor AP-2 alpha |
| chr23_+_36052944 | 0.32 |
ENSDART00000103149
|
hoxc13a
|
homeobox C13a |
| chr24_-_36271352 | 0.32 |
ENSDART00000153682
ENSDART00000155892 |
si:ch211-40k21.5
|
si:ch211-40k21.5 |
| chr11_-_13107106 | 0.31 |
ENSDART00000184477
|
elovl1b
|
ELOVL fatty acid elongase 1b |
| chr9_-_19583704 | 0.31 |
ENSDART00000053218
|
tmprss3a
|
transmembrane protease, serine 3a |
| chr21_+_5209476 | 0.31 |
ENSDART00000146400
|
st8sia5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5 |
| chr9_-_32847642 | 0.31 |
ENSDART00000121506
|
hpx
|
hemopexin |
| chr14_+_22172047 | 0.31 |
ENSDART00000114750
ENSDART00000148259 |
gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr19_-_25114701 | 0.31 |
ENSDART00000149035
|
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr9_+_43797902 | 0.30 |
ENSDART00000020550
|
ube2e3
|
ubiquitin-conjugating enzyme E2E 3 (UBC4/5 homolog, yeast) |
| chr7_+_27250186 | 0.30 |
ENSDART00000150068
|
sox6
|
SRY (sex determining region Y)-box 6 |
| chr20_+_36234335 | 0.30 |
ENSDART00000193484
ENSDART00000181664 |
cnih3
|
cornichon family AMPA receptor auxiliary protein 3 |
| chr7_+_50464500 | 0.30 |
ENSDART00000191356
|
serinc4
|
serine incorporator 4 |
| chr20_-_2134620 | 0.30 |
ENSDART00000064375
|
tmem244
|
transmembrane protein 244 |
| chr20_-_40451115 | 0.30 |
ENSDART00000075092
|
pkib
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr13_+_27298673 | 0.29 |
ENSDART00000142922
|
slc17a5
|
solute carrier family 17 (acidic sugar transporter), member 5 |
| chr8_+_38417461 | 0.29 |
ENSDART00000132718
|
nkx6.3
|
NK6 homeobox 3 |
| chr7_-_8738827 | 0.29 |
ENSDART00000172807
ENSDART00000173026 |
si:ch211-1o7.3
|
si:ch211-1o7.3 |
| chr24_-_8729716 | 0.29 |
ENSDART00000183624
|
tfap2a
|
transcription factor AP-2 alpha |
| chr1_+_51721851 | 0.29 |
ENSDART00000040397
|
prdx2
|
peroxiredoxin 2 |
| chr17_-_15029258 | 0.28 |
ENSDART00000168332
ENSDART00000012877 ENSDART00000109190 |
ero1a
|
endoplasmic reticulum oxidoreductase alpha |
| chr2_-_21349425 | 0.28 |
ENSDART00000171699
|
hhatla
|
hedgehog acyltransferase like, a |
| chr4_-_17741513 | 0.28 |
ENSDART00000141680
|
mybpc1
|
myosin binding protein C, slow type |
| chr12_-_47793857 | 0.27 |
ENSDART00000161294
|
dydc2
|
DPY30 domain containing 2 |
| chr13_+_12671513 | 0.26 |
ENSDART00000010517
|
eif4eb
|
eukaryotic translation initiation factor 4eb |
| chr21_-_22117085 | 0.26 |
ENSDART00000146673
|
slc35f2
|
solute carrier family 35, member F2 |
| chr19_-_3726768 | 0.26 |
ENSDART00000161738
|
smim13
|
small integral membrane protein 13 |
| chr24_-_39186185 | 0.25 |
ENSDART00000123019
ENSDART00000191114 |
nubp2
|
nucleotide binding protein 2 (MinD homolog, E. coli) |
| chr23_+_36460239 | 0.25 |
ENSDART00000172441
|
lima1a
|
LIM domain and actin binding 1a |
| chr13_-_48388726 | 0.25 |
ENSDART00000169473
|
fbxo11a
|
F-box protein 11a |
| chr2_+_2772447 | 0.25 |
ENSDART00000124882
|
thoc1
|
THO complex 1 |
| chr23_-_18381361 | 0.25 |
ENSDART00000016891
|
hsd17b10
|
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr1_+_40023640 | 0.25 |
ENSDART00000101623
|
lgi2b
|
leucine-rich repeat LGI family, member 2b |
| chr10_+_38560032 | 0.24 |
ENSDART00000177197
|
LO018336.1
|
|
| chr6_-_38418862 | 0.24 |
ENSDART00000104135
|
gabra5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| chr7_+_21887307 | 0.24 |
ENSDART00000052871
|
pop7
|
POP7 homolog, ribonuclease P/MRP subunit |
| chr4_-_13613148 | 0.23 |
ENSDART00000067164
ENSDART00000111247 |
irf5
|
interferon regulatory factor 5 |
| chr10_-_14556978 | 0.23 |
ENSDART00000126643
|
zgc:153395
|
zgc:153395 |
| chr4_+_3482312 | 0.23 |
ENSDART00000109044
|
grm8a
|
glutamate receptor, metabotropic 8a |
| chr15_-_5252673 | 0.23 |
ENSDART00000132963
|
or126-1
|
odorant receptor, family E, subfamily 126, member 1 |
| chr25_+_5015019 | 0.23 |
ENSDART00000127600
|
hdac10
|
histone deacetylase 10 |
| chr21_-_39566854 | 0.23 |
ENSDART00000020174
|
dynll2b
|
dynein, light chain, LC8-type 2b |
| chr9_-_1949915 | 0.23 |
ENSDART00000190712
|
hoxd3a
|
homeobox D3a |
| chr19_-_10306910 | 0.22 |
ENSDART00000151684
|
u2af2b
|
U2 small nuclear RNA auxiliary factor 2b |
| chr25_-_11057753 | 0.22 |
ENSDART00000186551
|
sv2bb
|
synaptic vesicle glycoprotein 2Bb |
| chr24_+_11908480 | 0.22 |
ENSDART00000024224
|
fen1
|
flap structure-specific endonuclease 1 |
| chr11_-_165288 | 0.22 |
ENSDART00000108703
ENSDART00000173151 |
tegt
|
testis enhanced gene transcript (BAX inhibitor 1) |
| chr3_+_55093581 | 0.22 |
ENSDART00000101709
|
hbaa2
|
hemoglobin, alpha adult 2 |
| chr14_-_12390724 | 0.22 |
ENSDART00000131343
|
magt1
|
magnesium transporter 1 |
| chr9_-_31915423 | 0.22 |
ENSDART00000060051
|
fgf14
|
fibroblast growth factor 14 |
| chr6_-_58757131 | 0.22 |
ENSDART00000083582
|
soat2
|
sterol O-acyltransferase 2 |
| chr22_-_36530902 | 0.22 |
ENSDART00000056188
|
polr2h
|
info polymerase (RNA) II (DNA directed) polypeptide H |
| chr11_-_3897067 | 0.21 |
ENSDART00000134858
|
rpn1
|
ribophorin I |
| chr14_+_29609245 | 0.21 |
ENSDART00000043058
|
TENM2
|
si:dkey-34l15.2 |
| chr22_-_881080 | 0.21 |
ENSDART00000185489
|
cept1b
|
choline/ethanolamine phosphotransferase 1b |
| chr1_+_5402476 | 0.21 |
ENSDART00000040204
|
tuba8l2
|
tubulin, alpha 8 like 2 |
| chr7_-_73843720 | 0.21 |
ENSDART00000111622
|
caap1
|
caspase activity and apoptosis inhibitor 1 |
| chr25_+_11008419 | 0.20 |
ENSDART00000156589
|
mhc1lia
|
major histocompatibility complex class I LIA |
| chr10_+_24690534 | 0.20 |
ENSDART00000079549
|
tpte
|
transmembrane phosphatase with tensin homology |
| chr7_-_17814118 | 0.20 |
ENSDART00000179688
|
ecsit
|
ECSIT signalling integrator |
| chr8_+_40628926 | 0.20 |
ENSDART00000163598
|
dusp2
|
dual specificity phosphatase 2 |
| chr14_+_6963312 | 0.20 |
ENSDART00000150050
|
hnrnpaba
|
heterogeneous nuclear ribonucleoprotein A/Ba |
| chr15_-_704408 | 0.20 |
ENSDART00000156200
ENSDART00000166404 ENSDART00000131040 |
zgc:174574
|
zgc:174574 |
| chr13_+_25455319 | 0.20 |
ENSDART00000145948
|
pkd2l1
|
polycystic kidney disease 2-like 1 |
| chr5_+_41477526 | 0.19 |
ENSDART00000153567
|
pias2
|
protein inhibitor of activated STAT, 2 |
| chr10_+_37500234 | 0.19 |
ENSDART00000132096
ENSDART00000099473 |
msi2a
|
musashi RNA-binding protein 2a |
| chr22_+_28337204 | 0.19 |
ENSDART00000163352
|
impg2b
|
interphotoreceptor matrix proteoglycan 2b |
| chr16_-_5105295 | 0.19 |
ENSDART00000082071
ENSDART00000148955 ENSDART00000184700 ENSDART00000188127 |
bckdhb
|
branched chain keto acid dehydrogenase E1, beta polypeptide |
| chr5_+_41477954 | 0.19 |
ENSDART00000185871
|
pias2
|
protein inhibitor of activated STAT, 2 |
| chr23_-_19230627 | 0.19 |
ENSDART00000007122
|
guca1b
|
guanylate cyclase activator 1B |
| chr25_-_8201983 | 0.18 |
ENSDART00000006579
|
saal1
|
serum amyloid A-like 1 |
| chr24_+_22021675 | 0.18 |
ENSDART00000081234
|
ropn1l
|
rhophilin associated tail protein 1-like |
| chr25_+_4635355 | 0.18 |
ENSDART00000021120
|
pgghg
|
protein-glucosylgalactosylhydroxylysine glucosidase |
| chr14_-_858985 | 0.18 |
ENSDART00000148687
ENSDART00000149375 |
slc34a1a
|
solute carrier family 34 (type II sodium/phosphate cotransporter), member 1a |
| chr24_+_28383278 | 0.18 |
ENSDART00000018095
|
sh3glb1a
|
SH3-domain GRB2-like endophilin B1a |
| chr14_+_31529958 | 0.18 |
ENSDART00000053026
|
fam122b
|
family with sequence similarity 122B |
| chr3_-_22366032 | 0.18 |
ENSDART00000029849
|
ifnphi1
|
interferon phi 1 |
| chr22_+_9091913 | 0.18 |
ENSDART00000186598
|
BX248395.1
|
|
| chr22_-_15385442 | 0.18 |
ENSDART00000090975
|
tmem264
|
transmembrane protein 264 |
| chr9_+_23003208 | 0.17 |
ENSDART00000021060
|
eaf2
|
ELL associated factor 2 |
| chr12_+_25845494 | 0.17 |
ENSDART00000170582
|
glud1b
|
glutamate dehydrogenase 1b |
| chr4_-_13931293 | 0.17 |
ENSDART00000067172
|
zcrb1
|
zinc finger CCHC-type and RNA binding motif 1 |
| chr7_+_24496894 | 0.17 |
ENSDART00000149994
|
nelfa
|
negative elongation factor complex member A |
| chr19_+_32201991 | 0.17 |
ENSDART00000022667
|
fam8a1a
|
family with sequence similarity 8, member A1a |
| chr6_-_37468971 | 0.17 |
ENSDART00000126379
|
plcxd1
|
phosphatidylinositol-specific phospholipase C, X domain containing 1 |
| chr4_-_13931508 | 0.17 |
ENSDART00000067174
|
zcrb1
|
zinc finger CCHC-type and RNA binding motif 1 |
| chr1_-_21409877 | 0.17 |
ENSDART00000102782
|
gria2a
|
glutamate receptor, ionotropic, AMPA 2a |
| chr3_-_58189429 | 0.16 |
ENSDART00000156092
|
si:ch211-256e16.11
|
si:ch211-256e16.11 |
| chr24_+_11908833 | 0.16 |
ENSDART00000178622
|
fen1
|
flap structure-specific endonuclease 1 |
| chr5_+_38684651 | 0.16 |
ENSDART00000137852
|
si:dkey-58f10.10
|
si:dkey-58f10.10 |
| chr4_+_74943111 | 0.16 |
ENSDART00000004739
|
nup50
|
nucleoporin 50 |
| chr5_-_47975440 | 0.16 |
ENSDART00000145665
ENSDART00000007057 |
ccnh
|
cyclin H |
| chr5_+_25072894 | 0.16 |
ENSDART00000012268
|
mrpl41
|
mitochondrial ribosomal protein L41 |
| chr5_-_26330313 | 0.16 |
ENSDART00000148656
|
arvcfb
|
ARVCF, delta catenin family member b |
| chr18_-_6151793 | 0.16 |
ENSDART00000122307
|
gcsha
|
glycine cleavage system protein H (aminomethyl carrier), a |
| chr11_+_30057762 | 0.16 |
ENSDART00000164139
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr22_-_3182965 | 0.16 |
ENSDART00000158009
|
lonp1
|
lon peptidase 1, mitochondrial |
| chr1_-_26045560 | 0.16 |
ENSDART00000172737
ENSDART00000076120 ENSDART00000193593 |
gpank1
|
G patch domain and ankyrin repeats 1 |
| chr20_-_25486384 | 0.15 |
ENSDART00000141340
|
si:dkey-183n20.15
|
si:dkey-183n20.15 |
| chr18_+_62932 | 0.15 |
ENSDART00000052638
|
slc27a2a
|
solute carrier family 27 (fatty acid transporter), member 2a |
| chr10_+_18557102 | 0.15 |
ENSDART00000160157
|
CR388008.1
|
|
| chr22_+_21398508 | 0.15 |
ENSDART00000089408
ENSDART00000186091 |
shdb
|
Src homology 2 domain containing transforming protein D, b |
| chr3_-_23513155 | 0.15 |
ENSDART00000170200
|
BX682558.1
|
|
| chr2_+_46032678 | 0.15 |
ENSDART00000184382
ENSDART00000125971 |
gpc1b
|
glypican 1b |
| chr7_+_48555626 | 0.15 |
ENSDART00000125483
ENSDART00000083514 |
kcnq1
|
potassium voltage-gated channel, KQT-like subfamily, member 1 |
| chr10_+_20392083 | 0.15 |
ENSDART00000166867
|
r3hcc1
|
R3H domain and coiled-coil containing 1 |
| chr5_+_38631119 | 0.15 |
ENSDART00000131832
ENSDART00000162215 |
si:ch211-271e10.6
|
si:ch211-271e10.6 |
| chr5_+_32490238 | 0.15 |
ENSDART00000191839
|
ndor1
|
NADPH dependent diflavin oxidoreductase 1 |
| chr1_+_29162478 | 0.15 |
ENSDART00000141441
|
ankrd10a
|
ankyrin repeat domain 10a |
| chr24_+_30695120 | 0.15 |
ENSDART00000178649
|
zgc:174719
|
zgc:174719 |
| chr9_+_23124252 | 0.15 |
ENSDART00000192377
|
BX936418.1
|
|
| chr5_+_26807142 | 0.15 |
ENSDART00000188981
|
rabl6b
|
RAB, member RAS oncogene family-like 6b |
| chr3_+_25914064 | 0.15 |
ENSDART00000162558
ENSDART00000166843 |
tom1
|
target of myb1 membrane trafficking protein |
| chr3_+_24189804 | 0.14 |
ENSDART00000134723
|
prr15la
|
proline rich 15-like a |
| chr15_+_9121054 | 0.14 |
ENSDART00000193889
|
ptgir
|
prostaglandin I2 receptor |
| chr22_+_29135846 | 0.14 |
ENSDART00000143811
|
baiap2l2b
|
BAI1-associated protein 2-like 2b |
| chr14_+_8343498 | 0.14 |
ENSDART00000164551
|
nrg2b
|
neuregulin 2b |
| chr11_-_40728380 | 0.14 |
ENSDART00000023745
|
ccdc114
|
coiled-coil domain containing 114 |
| chr14_+_30762131 | 0.14 |
ENSDART00000145039
|
si:ch211-145o7.3
|
si:ch211-145o7.3 |
| chr9_+_8365398 | 0.14 |
ENSDART00000138713
ENSDART00000136847 |
si:dkey-90l23.2
|
si:dkey-90l23.2 |
| chr7_+_45975537 | 0.14 |
ENSDART00000170253
|
plekhf1
|
pleckstrin homology domain containing, family F (with FYVE domain) member 1 |
| chr19_-_11238620 | 0.14 |
ENSDART00000148697
ENSDART00000149169 |
ssr2
|
signal sequence receptor, beta |
| chr14_+_33507608 | 0.14 |
ENSDART00000038128
|
mcts1
|
malignant T cell amplified sequence 1 |
| chr15_-_1001177 | 0.14 |
ENSDART00000160730
|
zgc:162936
|
zgc:162936 |
| chr13_-_39159810 | 0.14 |
ENSDART00000131508
|
col9a1b
|
collagen, type IX, alpha 1b |
| chr15_-_25319663 | 0.14 |
ENSDART00000042218
|
pafah1b1a
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1a |
| chr2_-_48753873 | 0.14 |
ENSDART00000189556
|
CABZ01044731.1
|
|
| chr2_+_25657958 | 0.14 |
ENSDART00000161407
|
tnika
|
TRAF2 and NCK interacting kinase a |
Network of associatons between targets according to the STRING database.
First level regulatory network of pax10+pax6a+pax6b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.2 | 0.5 | GO:0046099 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine biosynthetic process(GO:0046099) |
| 0.1 | 0.4 | GO:0050995 | negative regulation of lipid catabolic process(GO:0050995) |
| 0.1 | 0.7 | GO:0021767 | mammillary body development(GO:0021767) |
| 0.1 | 0.3 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 0.1 | 0.8 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 0.3 | GO:0060254 | N-terminal protein palmitoylation(GO:0006500) negative regulation of lipoprotein metabolic process(GO:0050748) regulation of N-terminal protein palmitoylation(GO:0060254) negative regulation of N-terminal protein palmitoylation(GO:0060262) negative regulation of protein lipidation(GO:1903060) |
| 0.1 | 0.3 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.1 | 0.5 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.1 | 0.4 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 1.0 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.1 | 0.4 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.4 | GO:0043090 | amino acid import(GO:0043090) L-amino acid import(GO:0043092) |
| 0.0 | 0.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.2 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.1 | GO:0048662 | regulation of smooth muscle cell proliferation(GO:0048660) negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.0 | 0.3 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.6 | GO:0014034 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.2 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:0051230 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.0 | 0.2 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.7 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) semicircular canal fusion(GO:0060879) |
| 0.0 | 0.7 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.2 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.0 | 0.3 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.3 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.3 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.1 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.2 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.3 | GO:0071450 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.5 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.7 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.1 | GO:1904251 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.0 | 0.2 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.0 | 0.4 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.2 | GO:0034435 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.0 | 0.2 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.2 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.0 | 0.1 | GO:0007508 | larval development(GO:0002164) larval heart development(GO:0007508) |
| 0.0 | 0.1 | GO:0010359 | regulation of anion channel activity(GO:0010359) |
| 0.0 | 1.0 | GO:0046883 | regulation of hormone secretion(GO:0046883) |
| 0.0 | 0.1 | GO:0071548 | response to dexamethasone(GO:0071548) |
| 0.0 | 0.5 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.0 | 0.4 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.4 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 0.1 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.8 | GO:0045103 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.1 | GO:0045337 | farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.0 | 0.4 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:0006203 | dGTP catabolic process(GO:0006203) purine nucleoside triphosphate catabolic process(GO:0009146) purine deoxyribonucleoside triphosphate catabolic process(GO:0009217) dGTP metabolic process(GO:0046070) |
| 0.0 | 0.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.2 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.1 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 0.4 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.1 | GO:0016139 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.0 | 0.2 | GO:0032785 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.4 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.2 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.1 | GO:0034163 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.3 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.1 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.4 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.1 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.1 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.2 | GO:0014812 | muscle cell migration(GO:0014812) |
| 0.0 | 0.2 | GO:0042744 | oxygen transport(GO:0015671) hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.2 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.0 | 0.2 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.1 | GO:0044090 | positive regulation of vacuole organization(GO:0044090) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 0.8 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.2 | GO:0030062 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.1 | 0.2 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.0 | 0.1 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.3 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.4 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.1 | GO:0008247 | 1-alkyl-2-acetylglycerophosphocholine esterase complex(GO:0008247) |
| 0.0 | 0.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.4 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.2 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.2 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.2 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 1.0 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.1 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.5 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.2 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.5 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 1.2 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.2 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.0 | 0.1 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.2 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0047777 | (3S)-citramalyl-CoA lyase activity(GO:0047777) |
| 0.2 | 0.5 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.1 | 4.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 0.5 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 0.8 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.4 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 0.7 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.1 | 0.4 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.1 | 0.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 0.2 | GO:0047611 | acetylspermidine deacetylase activity(GO:0047611) |
| 0.1 | 0.7 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 0.4 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 0.2 | GO:0034736 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.0 | 0.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.4 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.3 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.2 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.1 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 0.3 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.2 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.2 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 0.1 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.7 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.1 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.0 | 0.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.4 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.2 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.4 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.2 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.2 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.2 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.2 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.3 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.5 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.1 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.5 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 0.2 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.1 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.2 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.3 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.4 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.2 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.3 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 1.3 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.0 | 0.2 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.3 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.2 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.8 | GO:1902936 | phosphatidylinositol bisphosphate binding(GO:1902936) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.3 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.4 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.2 | PID CONE PATHWAY | Visual signal transduction: Cones |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 1.1 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.4 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 0.0 | 0.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.5 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.1 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |