Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
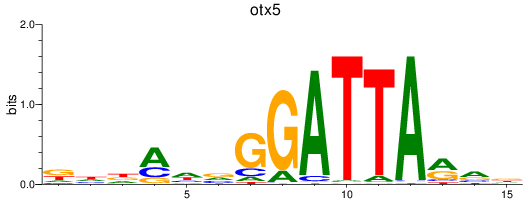
Results for otx5
Z-value: 0.85
Transcription factors associated with otx5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
otx5
|
ENSDARG00000043483 | orthodenticle homolog 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| otx5 | dr11_v1_chr15_+_47903864_47903864 | -0.27 | 2.7e-01 | Click! |
Activity profile of otx5 motif
Sorted Z-values of otx5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_25737452 | 1.70 |
ENSDART00000152021
|
si:ch211-214p16.3
|
si:ch211-214p16.3 |
| chr4_+_18804317 | 1.64 |
ENSDART00000101043
|
slc26a3.2
|
solute carrier family 26 (anion exchanger), member 3, tandem duplicate 2 |
| chr18_-_40684756 | 1.46 |
ENSDART00000113799
ENSDART00000139042 |
si:ch211-132b12.7
|
si:ch211-132b12.7 |
| chr13_+_23176330 | 1.44 |
ENSDART00000168351
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr19_-_2115040 | 1.31 |
ENSDART00000020497
|
snx13
|
sorting nexin 13 |
| chr17_+_50701748 | 1.10 |
ENSDART00000191938
ENSDART00000183220 ENSDART00000049464 |
fermt2
|
fermitin family member 2 |
| chr15_-_5815006 | 1.03 |
ENSDART00000102459
|
rbp2a
|
retinol binding protein 2a, cellular |
| chr2_-_48171112 | 1.02 |
ENSDART00000156258
|
pfkpb
|
phosphofructokinase, platelet b |
| chr3_-_49138004 | 0.99 |
ENSDART00000167173
|
gipc1
|
GIPC PDZ domain containing family, member 1 |
| chr23_+_39963599 | 0.98 |
ENSDART00000166539
|
fyco1a
|
FYVE and coiled-coil domain containing 1a |
| chr17_-_6730247 | 0.98 |
ENSDART00000031091
|
vsnl1b
|
visinin-like 1b |
| chr4_-_8030583 | 0.97 |
ENSDART00000113628
|
si:ch211-240l19.8
|
si:ch211-240l19.8 |
| chr25_-_17552299 | 0.95 |
ENSDART00000154327
|
si:dkey-44k1.5
|
si:dkey-44k1.5 |
| chr1_-_58009216 | 0.95 |
ENSDART00000143829
|
nxnl1
|
nucleoredoxin like 1 |
| chr11_+_25583950 | 0.94 |
ENSDART00000111961
|
ccdc120
|
coiled-coil domain containing 120 |
| chr2_+_7192966 | 0.94 |
ENSDART00000142735
|
si:ch211-13f8.1
|
si:ch211-13f8.1 |
| chr23_+_39854566 | 0.92 |
ENSDART00000190423
ENSDART00000164473 ENSDART00000161881 |
si:ch73-217b7.1
|
si:ch73-217b7.1 |
| chr23_-_40250886 | 0.92 |
ENSDART00000041912
|
tgm1l3
|
transglutaminase 1 like 3 |
| chr8_+_2656231 | 0.92 |
ENSDART00000160833
|
fam102aa
|
family with sequence similarity 102, member Aa |
| chr1_-_53407448 | 0.91 |
ENSDART00000160033
ENSDART00000172322 |
elmod2
|
ELMO/CED-12 domain containing 2 |
| chr10_+_32683089 | 0.91 |
ENSDART00000063551
|
ppm1e
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E |
| chr15_-_47115787 | 0.90 |
ENSDART00000192601
|
CABZ01079081.1
|
|
| chr9_-_27442339 | 0.86 |
ENSDART00000138602
|
stxbp5l
|
syntaxin binding protein 5-like |
| chr7_-_72208248 | 0.85 |
ENSDART00000108916
|
zmp:0000001168
|
zmp:0000001168 |
| chr3_+_49521106 | 0.83 |
ENSDART00000162799
|
crb3a
|
crumbs homolog 3a |
| chr18_+_2189211 | 0.82 |
ENSDART00000170827
|
ccpg1
|
cell cycle progression 1 |
| chr3_+_3610140 | 0.81 |
ENSDART00000104977
|
pick1
|
protein interacting with prkca 1 |
| chr11_+_6152643 | 0.81 |
ENSDART00000012789
|
slc1a6
|
solute carrier family 1 (high affinity aspartate/glutamate transporter), member 6 |
| chr1_-_46859398 | 0.79 |
ENSDART00000135795
|
agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr2_+_51783120 | 0.77 |
ENSDART00000177559
|
crygn1
|
crystallin, gamma N1 |
| chr8_+_36560019 | 0.77 |
ENSDART00000136418
ENSDART00000061378 ENSDART00000185237 |
sf3a1
|
splicing factor 3a, subunit 1 |
| chr20_+_29587995 | 0.75 |
ENSDART00000153339
|
adam17b
|
ADAM metallopeptidase domain 17b |
| chr3_+_12725343 | 0.73 |
ENSDART00000188583
|
cyp2k20
|
cytochrome P450, family 2, subfamily k, polypeptide 20 |
| chr14_+_15620640 | 0.70 |
ENSDART00000188867
|
si:dkey-203a12.9
|
si:dkey-203a12.9 |
| chr9_-_12574473 | 0.69 |
ENSDART00000191372
ENSDART00000193667 |
igf2bp2a
|
insulin-like growth factor 2 mRNA binding protein 2a |
| chr19_-_9503473 | 0.69 |
ENSDART00000091615
|
iffo1a
|
intermediate filament family orphan 1a |
| chr1_-_11075403 | 0.68 |
ENSDART00000102903
ENSDART00000170290 |
dmd
|
dystrophin |
| chr21_-_3770636 | 0.64 |
ENSDART00000053596
|
scamp1
|
secretory carrier membrane protein 1 |
| chr7_-_6592142 | 0.64 |
ENSDART00000160137
|
kcnj10a
|
potassium inwardly-rectifying channel, subfamily J, member 10a |
| chr8_-_5850472 | 0.64 |
ENSDART00000179217
|
CABZ01102147.1
|
|
| chr10_+_28428222 | 0.63 |
ENSDART00000135003
|
si:ch211-222e20.4
|
si:ch211-222e20.4 |
| chr9_+_13685921 | 0.63 |
ENSDART00000145775
|
mpp4a
|
membrane protein, palmitoylated 4a (MAGUK p55 subfamily member 4) |
| chr4_-_77432218 | 0.62 |
ENSDART00000158683
|
slco1d1
|
solute carrier organic anion transporter family, member 1D1 |
| chr24_+_37723362 | 0.62 |
ENSDART00000136836
|
rab11fip3
|
RAB11 family interacting protein 3 (class II) |
| chr23_+_39558508 | 0.62 |
ENSDART00000017902
|
camk1gb
|
calcium/calmodulin-dependent protein kinase IGb |
| chr20_-_33566640 | 0.61 |
ENSDART00000159729
|
si:dkey-65b13.9
|
si:dkey-65b13.9 |
| chr4_-_47096170 | 0.61 |
ENSDART00000161079
|
si:dkey-26m3.3
|
si:dkey-26m3.3 |
| chr14_+_26796917 | 0.59 |
ENSDART00000137695
|
klhl4
|
kelch-like family member 4 |
| chr2_-_48171441 | 0.58 |
ENSDART00000123040
|
pfkpb
|
phosphofructokinase, platelet b |
| chr3_+_16762483 | 0.57 |
ENSDART00000132732
|
tmem86b
|
transmembrane protein 86B |
| chr17_-_42492668 | 0.56 |
ENSDART00000183946
|
CR925755.2
|
|
| chr19_-_9760994 | 0.54 |
ENSDART00000139727
|
si:dkey-14o18.2
|
si:dkey-14o18.2 |
| chr24_-_38079261 | 0.53 |
ENSDART00000105662
|
crp1
|
C-reactive protein 1 |
| chr22_-_8509215 | 0.53 |
ENSDART00000140146
|
si:ch73-27e22.3
|
si:ch73-27e22.3 |
| chr23_+_16935494 | 0.53 |
ENSDART00000143120
|
si:dkey-147f3.4
|
si:dkey-147f3.4 |
| chr14_+_2487672 | 0.52 |
ENSDART00000170629
ENSDART00000123063 |
fgf18a
|
fibroblast growth factor 18a |
| chr8_-_50979047 | 0.51 |
ENSDART00000184788
ENSDART00000180906 |
zgc:91909
|
zgc:91909 |
| chr4_-_75651505 | 0.50 |
ENSDART00000115143
|
si:dkey-71l4.2
|
si:dkey-71l4.2 |
| chr3_+_48561112 | 0.50 |
ENSDART00000159682
|
slc16a5b
|
solute carrier family 16 (monocarboxylate transporter), member 5b |
| chr4_+_75575252 | 0.50 |
ENSDART00000166536
|
si:ch211-227e10.1
|
si:ch211-227e10.1 |
| chr5_-_1827575 | 0.50 |
ENSDART00000171193
|
mcc
|
mutated in colorectal cancers |
| chr4_-_76303301 | 0.49 |
ENSDART00000158504
|
zgc:174944
|
zgc:174944 |
| chr3_-_55404985 | 0.48 |
ENSDART00000154274
|
mafga
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Ga |
| chr17_+_35097024 | 0.48 |
ENSDART00000026152
|
asap2a
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2a |
| chr4_+_65487457 | 0.47 |
ENSDART00000162017
ENSDART00000193241 |
si:dkeyp-5g9.1
|
si:dkeyp-5g9.1 |
| chr6_+_46481024 | 0.46 |
ENSDART00000155596
|
si:dkey-7k24.5
|
si:dkey-7k24.5 |
| chr21_+_11105007 | 0.45 |
ENSDART00000128859
ENSDART00000193105 |
prlra
|
prolactin receptor a |
| chr14_-_2318590 | 0.45 |
ENSDART00000192735
|
pcdh2ab8
|
protocadherin 2 alpha b 8 |
| chr4_-_63558838 | 0.45 |
ENSDART00000191708
ENSDART00000190740 ENSDART00000186476 |
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr6_-_15604417 | 0.45 |
ENSDART00000157817
|
lrrfip1b
|
leucine rich repeat (in FLII) interacting protein 1b |
| chr14_+_3038473 | 0.44 |
ENSDART00000026021
ENSDART00000150000 |
cd74a
|
CD74 molecule, major histocompatibility complex, class II invariant chain a |
| chr10_+_40204175 | 0.44 |
ENSDART00000188277
|
gramd1ba
|
GRAM domain containing 1Ba |
| chr1_+_55080797 | 0.44 |
ENSDART00000077390
|
lgalsla
|
lectin, galactoside-binding-like a |
| chr22_+_35516440 | 0.44 |
ENSDART00000184191
|
hykk.2
|
hydroxylysine kinase, tandem duplicate 2 |
| chr1_+_45217425 | 0.44 |
ENSDART00000179983
ENSDART00000074683 |
EVI5L
|
si:ch211-239f4.1 |
| chr4_-_71267777 | 0.43 |
ENSDART00000169664
|
si:dkeyp-51g9.4
|
si:dkeyp-51g9.4 |
| chr4_-_33073382 | 0.43 |
ENSDART00000146414
|
si:dkeyp-4f2.1
|
si:dkeyp-4f2.1 |
| chr3_-_58165254 | 0.43 |
ENSDART00000093031
|
snu13a
|
SNU13 homolog, small nuclear ribonucleoprotein a (U4/U6.U5) |
| chr4_+_58661331 | 0.42 |
ENSDART00000188763
|
CR388148.2
|
|
| chr16_+_34111919 | 0.41 |
ENSDART00000134037
ENSDART00000006061 ENSDART00000140552 |
tcea3
|
transcription elongation factor A (SII), 3 |
| chr7_-_17780048 | 0.41 |
ENSDART00000183336
|
si:dkey-106g10.7
|
si:dkey-106g10.7 |
| chr11_-_3629201 | 0.41 |
ENSDART00000136577
ENSDART00000132121 |
itih3a
|
inter-alpha-trypsin inhibitor heavy chain 3a |
| chr17_+_25677458 | 0.41 |
ENSDART00000029703
|
kcnh1a
|
potassium voltage-gated channel, subfamily H (eag-related), member 1a |
| chr4_+_45389591 | 0.40 |
ENSDART00000162530
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr17_-_7371564 | 0.40 |
ENSDART00000060336
|
rab32b
|
RAB32b, member RAS oncogene family |
| chr5_+_31791001 | 0.40 |
ENSDART00000043010
|
slc25a25b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25b |
| chr15_-_29510074 | 0.40 |
ENSDART00000184070
|
BX324227.3
|
|
| chr8_+_54263946 | 0.40 |
ENSDART00000193119
|
TMCC1 (1 of many)
|
transmembrane and coiled-coil domain family 1 |
| chr4_-_38828508 | 0.39 |
ENSDART00000151952
|
si:dkey-59l11.10
|
si:dkey-59l11.10 |
| chr6_+_20650003 | 0.39 |
ENSDART00000049465
|
slc19a1
|
solute carrier family 19 (folate transporter), member 1 |
| chr10_-_20445549 | 0.38 |
ENSDART00000064613
|
loxl2a
|
lysyl oxidase-like 2a |
| chr13_-_14269626 | 0.37 |
ENSDART00000079176
|
gfra4a
|
GDNF family receptor alpha 4a |
| chr6_-_59381391 | 0.37 |
ENSDART00000157066
|
ppp1r16b
|
protein phosphatase 1, regulatory subunit 16B |
| chr3_+_26144765 | 0.36 |
ENSDART00000146267
ENSDART00000043932 |
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr10_+_39476432 | 0.36 |
ENSDART00000190375
ENSDART00000183954 |
kirrel3a
|
kirre like nephrin family adhesion molecule 3a |
| chr4_+_58667348 | 0.36 |
ENSDART00000186596
ENSDART00000180673 |
CR388148.2
|
|
| chr16_-_40043322 | 0.36 |
ENSDART00000145278
|
si:dkey-29b11.3
|
si:dkey-29b11.3 |
| chr5_-_69180587 | 0.35 |
ENSDART00000156681
ENSDART00000160753 |
zgc:171967
|
zgc:171967 |
| chr19_+_42694211 | 0.35 |
ENSDART00000180153
|
clasp2
|
cytoplasmic linker associated protein 2 |
| chr4_+_49664256 | 0.34 |
ENSDART00000155251
|
znf974
|
zinc finger protein 974 |
| chr25_-_34632050 | 0.34 |
ENSDART00000170671
|
megf8
|
multiple EGF-like-domains 8 |
| chr4_+_65271779 | 0.33 |
ENSDART00000134257
|
znf1027
|
zinc finger protein 1027 |
| chr2_+_30878864 | 0.33 |
ENSDART00000009326
|
oprk1
|
opioid receptor, kappa 1 |
| chr4_+_35227135 | 0.33 |
ENSDART00000161023
ENSDART00000182403 |
si:ch211-276i6.1
|
si:ch211-276i6.1 |
| chr18_-_42071876 | 0.33 |
ENSDART00000193288
ENSDART00000189021 ENSDART00000019245 |
arhgap42b
|
Rho GTPase activating protein 42b |
| chr15_+_40079468 | 0.33 |
ENSDART00000154947
|
ngef
|
neuronal guanine nucleotide exchange factor |
| chr4_-_67984478 | 0.33 |
ENSDART00000160623
|
si:ch211-223k15.1
|
si:ch211-223k15.1 |
| chr4_-_52783184 | 0.32 |
ENSDART00000172283
|
si:dkey-4j21.2
|
si:dkey-4j21.2 |
| chr13_+_1015749 | 0.32 |
ENSDART00000190982
|
prokr1b
|
prokineticin receptor 1b |
| chr17_+_4578552 | 0.32 |
ENSDART00000038730
ENSDART00000182146 |
zmp:0000000619
|
zmp:0000000619 |
| chr10_-_26218354 | 0.32 |
ENSDART00000180764
|
arfip2b
|
ADP-ribosylation factor interacting protein 2b |
| chr3_-_18274691 | 0.32 |
ENSDART00000161140
|
BX649434.3
|
|
| chr4_-_76105527 | 0.32 |
ENSDART00000159108
|
zgc:110171
|
zgc:110171 |
| chr10_-_36738619 | 0.31 |
ENSDART00000093000
ENSDART00000157179 |
PLEKHB1
|
si:ch211-176g13.7 |
| chr23_-_7797207 | 0.31 |
ENSDART00000181611
|
myt1b
|
myelin transcription factor 1b |
| chr18_+_6641542 | 0.31 |
ENSDART00000160379
|
c2cd5
|
C2 calcium-dependent domain containing 5 |
| chr15_-_568645 | 0.31 |
ENSDART00000156744
|
cbln18
|
cerebellin 18 |
| chr7_-_59515569 | 0.30 |
ENSDART00000163343
ENSDART00000165457 ENSDART00000163745 |
slx1b
|
SLX1 homolog B, structure-specific endonuclease subunit |
| chr9_+_1313418 | 0.29 |
ENSDART00000191361
|
casp8l2
|
caspase 8, apoptosis-related cysteine peptidase, like 2 |
| chr1_+_14348399 | 0.29 |
ENSDART00000064468
|
crmp1
|
collapsin response mediator protein 1 |
| chr22_+_38164486 | 0.29 |
ENSDART00000137521
|
tm4sf18
|
transmembrane 4 L six family member 18 |
| chr4_-_70263609 | 0.29 |
ENSDART00000163868
|
si:dkey-3h2.3
|
si:dkey-3h2.3 |
| chr4_+_52177103 | 0.29 |
ENSDART00000166189
|
si:dkey-16b10.1
|
si:dkey-16b10.1 |
| chr7_-_48173440 | 0.29 |
ENSDART00000124075
|
mtss1lb
|
metastasis suppressor 1-like b |
| chr5_-_30074332 | 0.28 |
ENSDART00000147963
|
bco2a
|
beta-carotene oxygenase 2a |
| chr5_+_20148671 | 0.28 |
ENSDART00000143205
|
svopa
|
SV2 related protein a |
| chr25_+_192116 | 0.28 |
ENSDART00000153983
|
zgc:114188
|
zgc:114188 |
| chr4_-_63559311 | 0.28 |
ENSDART00000164415
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr4_-_71214516 | 0.27 |
ENSDART00000169997
|
si:ch211-205a14.7
|
si:ch211-205a14.7 |
| chr4_+_45388995 | 0.27 |
ENSDART00000150802
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr4_-_43185261 | 0.27 |
ENSDART00000150380
|
si:dkey-16p6.2
|
si:dkey-16p6.2 |
| chr23_+_7505943 | 0.27 |
ENSDART00000135787
|
hck
|
HCK proto-oncogene, Src family tyrosine kinase |
| chr9_+_20519846 | 0.26 |
ENSDART00000109680
ENSDART00000142220 |
vtcn1
|
V-set domain containing T cell activation inhibitor 1 |
| chr20_-_28931901 | 0.26 |
ENSDART00000153082
ENSDART00000046042 |
susd6
|
sushi domain containing 6 |
| chr11_+_29671661 | 0.25 |
ENSDART00000024318
ENSDART00000165024 |
rnf207a
|
ring finger protein 207a |
| chr4_-_75795899 | 0.25 |
ENSDART00000157925
|
si:dkey-261j11.1
|
si:dkey-261j11.1 |
| chr23_-_31266586 | 0.25 |
ENSDART00000139746
|
si:dkey-261l7.2
|
si:dkey-261l7.2 |
| chr4_-_38829098 | 0.24 |
ENSDART00000151932
|
si:dkey-59l11.10
|
si:dkey-59l11.10 |
| chr3_+_832788 | 0.24 |
ENSDART00000193025
|
FP102888.2
|
|
| chr3_+_35608385 | 0.24 |
ENSDART00000193219
ENSDART00000132703 |
traf7
|
TNF receptor-associated factor 7 |
| chr16_+_52343905 | 0.24 |
ENSDART00000131051
|
ifnlr1
|
interferon lambda receptor 1 |
| chr7_-_65114067 | 0.24 |
ENSDART00000110614
ENSDART00000098277 |
tmem41b
|
transmembrane protein 41B |
| chr3_+_59851537 | 0.24 |
ENSDART00000180997
|
CU693479.1
|
|
| chr15_-_40065265 | 0.24 |
ENSDART00000063781
ENSDART00000142394 |
gpr55a
|
G protein-coupled receptor 55a |
| chr4_+_33818711 | 0.23 |
ENSDART00000186452
|
znf1026
|
zinc finger protein 1026 |
| chr5_-_67145505 | 0.23 |
ENSDART00000011295
|
rom1a
|
retinal outer segment membrane protein 1a |
| chr4_-_49133107 | 0.23 |
ENSDART00000150806
|
znf1146
|
zinc finger protein 1146 |
| chr6_+_32834760 | 0.23 |
ENSDART00000121562
|
cyldl
|
cylindromatosis (turban tumor syndrome), like |
| chr21_-_42007213 | 0.23 |
ENSDART00000188804
ENSDART00000092821 ENSDART00000165743 |
gabrg2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr4_-_38539463 | 0.23 |
ENSDART00000158211
|
si:ch211-209n20.3
|
si:ch211-209n20.3 |
| chr4_+_51201875 | 0.22 |
ENSDART00000189875
ENSDART00000186722 |
BX005380.1
|
|
| chr4_-_4387012 | 0.22 |
ENSDART00000191836
|
CU468826.3
|
Danio rerio U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit-related protein 1-like (LOC100331497), mRNA. |
| chr17_-_51893123 | 0.22 |
ENSDART00000103350
ENSDART00000017329 |
numb
|
numb homolog (Drosophila) |
| chr12_+_3912544 | 0.22 |
ENSDART00000013465
|
tbx6
|
T-box 6 |
| chr4_-_43919994 | 0.22 |
ENSDART00000150297
|
znf1104
|
zinc finger protein 1104 |
| chr4_+_25558849 | 0.22 |
ENSDART00000113663
ENSDART00000100755 ENSDART00000111416 ENSDART00000127840 ENSDART00000168618 ENSDART00000111820 ENSDART00000113866 ENSDART00000110107 ENSDART00000111344 ENSDART00000108548 |
zgc:195175
|
zgc:195175 |
| chr4_-_76186742 | 0.22 |
ENSDART00000163970
|
si:ch211-106j21.4
|
si:ch211-106j21.4 |
| chr4_-_71416263 | 0.22 |
ENSDART00000159812
|
si:ch211-76m11.15
|
si:ch211-76m11.15 |
| chr19_+_14921000 | 0.21 |
ENSDART00000144052
|
oprd1a
|
opioid receptor, delta 1a |
| chr14_+_15430991 | 0.21 |
ENSDART00000158221
|
si:dkey-203a12.5
|
si:dkey-203a12.5 |
| chr5_-_69180227 | 0.21 |
ENSDART00000154816
|
zgc:171967
|
zgc:171967 |
| chr4_-_33763221 | 0.21 |
ENSDART00000132613
ENSDART00000191816 ENSDART00000188473 |
si:ch211-197f20.1
|
si:ch211-197f20.1 |
| chr1_+_8111009 | 0.21 |
ENSDART00000152192
|
SLC5A10
|
si:dkeyp-9d4.5 |
| chr9_-_3519253 | 0.21 |
ENSDART00000169586
|
dcaf17
|
ddb1 and cul4 associated factor 17 |
| chr19_-_35340027 | 0.21 |
ENSDART00000146394
|
macf1a
|
microtubule-actin crosslinking factor 1a |
| chr4_+_71014655 | 0.21 |
ENSDART00000170837
|
si:dkeyp-80d11.14
|
si:dkeyp-80d11.14 |
| chr4_-_56103062 | 0.20 |
ENSDART00000165784
ENSDART00000190846 |
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr1_+_8110562 | 0.20 |
ENSDART00000112160
|
SLC5A10
|
si:dkeyp-9d4.5 |
| chr4_+_60428812 | 0.20 |
ENSDART00000162049
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr4_+_64755464 | 0.20 |
ENSDART00000129549
|
znf1018
|
zinc finger protein 1018 |
| chr11_-_3366782 | 0.20 |
ENSDART00000127982
|
suox
|
sulfite oxidase |
| chr4_+_67994238 | 0.19 |
ENSDART00000160968
|
si:ch211-133h13.1
|
si:ch211-133h13.1 |
| chr4_+_43245188 | 0.19 |
ENSDART00000150310
|
si:dkey-29j8.1
|
si:dkey-29j8.1 |
| chr3_+_9071806 | 0.19 |
ENSDART00000162372
|
znf985
|
zinc finger protein 985 |
| chr16_+_52999778 | 0.18 |
ENSDART00000011506
|
nkd2a
|
naked cuticle homolog 2a |
| chr15_-_19669770 | 0.18 |
ENSDART00000152707
|
si:dkey-4p15.3
|
si:dkey-4p15.3 |
| chr4_-_51045333 | 0.18 |
ENSDART00000150560
|
znf1026
|
zinc finger protein 1026 |
| chr4_-_63769637 | 0.18 |
ENSDART00000158757
|
znf1098
|
zinc finger protein 1098 |
| chr4_+_69712223 | 0.18 |
ENSDART00000163035
ENSDART00000164475 |
si:dkey-238o14.1
zgc:174944
|
si:dkey-238o14.1 zgc:174944 |
| chr4_+_76973771 | 0.18 |
ENSDART00000142978
|
si:dkey-240n22.3
|
si:dkey-240n22.3 |
| chr13_+_22863516 | 0.18 |
ENSDART00000113082
ENSDART00000189200 |
hnrnph3
|
heterogeneous nuclear ribonucleoprotein H3 (2H9) |
| chr4_+_62431132 | 0.18 |
ENSDART00000160226
|
si:ch211-79g12.1
|
si:ch211-79g12.1 |
| chr8_-_43574935 | 0.18 |
ENSDART00000051139
|
ncor2
|
nuclear receptor corepressor 2 |
| chr4_+_47782411 | 0.17 |
ENSDART00000161728
|
si:ch211-196h24.2
|
si:ch211-196h24.2 |
| chr4_+_43225728 | 0.17 |
ENSDART00000158639
ENSDART00000141339 ENSDART00000137770 ENSDART00000166019 |
si:ch211-76m11.5
|
si:ch211-76m11.5 |
| chr4_-_71471302 | 0.16 |
ENSDART00000187687
|
si:ch211-76m11.5
|
si:ch211-76m11.5 |
| chr1_-_633356 | 0.16 |
ENSDART00000171019
|
appa
|
amyloid beta (A4) precursor protein a |
| chr15_+_5973909 | 0.16 |
ENSDART00000126886
ENSDART00000189618 |
igsf5b
|
immunoglobulin superfamily, member 5b |
| chr4_-_43919359 | 0.16 |
ENSDART00000184163
ENSDART00000158019 ENSDART00000191745 ENSDART00000181638 ENSDART00000150827 |
znf1104
|
zinc finger protein 1104 |
| chr15_+_45640906 | 0.16 |
ENSDART00000149361
ENSDART00000149079 |
sagb
|
S-antigen; retina and pineal gland (arrestin) b |
| chr7_-_12789251 | 0.15 |
ENSDART00000052750
|
adamtsl3
|
ADAMTS-like 3 |
| chr20_-_25412181 | 0.15 |
ENSDART00000062780
|
itsn2a
|
intersectin 2a |
| chr9_+_3519191 | 0.15 |
ENSDART00000008606
|
mettl8
|
methyltransferase like 8 |
| chr4_+_70341246 | 0.15 |
ENSDART00000123408
|
si:ch211-76m11.3
|
si:ch211-76m11.3 |
| chr14_+_43554490 | 0.15 |
ENSDART00000060351
|
adra1bb
|
adrenoceptor alpha 1Bb |
| chr17_-_8592824 | 0.14 |
ENSDART00000127022
|
CU462878.1
|
|
| chr8_+_23391263 | 0.14 |
ENSDART00000149687
|
si:dkey-16n15.6
|
si:dkey-16n15.6 |
| chr4_-_39186893 | 0.14 |
ENSDART00000150371
ENSDART00000121804 ENSDART00000133901 |
si:ch211-22k7.9
|
si:ch211-22k7.9 |
| chr21_-_33126697 | 0.14 |
ENSDART00000189293
ENSDART00000084559 |
CU019662.1
|
|
| chr4_-_47095583 | 0.14 |
ENSDART00000183595
ENSDART00000183279 ENSDART00000191638 |
si:dkey-26m3.3
|
si:dkey-26m3.3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of otx5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0010658 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.4 | 1.5 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.2 | 1.0 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.1 | 0.4 | GO:0010935 | macrophage cytokine production(GO:0010934) regulation of macrophage cytokine production(GO:0010935) positive regulation of macrophage cytokine production(GO:0060907) negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.1 | 0.5 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.1 | 0.8 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) |
| 0.1 | 0.4 | GO:0098838 | folic acid transport(GO:0015884) drug transport(GO:0015893) methotrexate transport(GO:0051958) reduced folate transmembrane transport(GO:0098838) |
| 0.1 | 0.8 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.5 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 1.0 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.1 | 0.4 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.1 | 0.6 | GO:0036268 | swimming(GO:0036268) |
| 0.1 | 0.4 | GO:0090076 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 0.1 | 0.6 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 0.9 | GO:0061718 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 0.3 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.1 | 1.6 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.5 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 | 0.3 | GO:0010828 | positive regulation of glucose transport(GO:0010828) |
| 0.1 | 0.7 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.2 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:1903441 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.0 | 0.9 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.4 | GO:0021576 | hindbrain formation(GO:0021576) |
| 0.0 | 0.8 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.8 | GO:1901888 | regulation of cell junction assembly(GO:1901888) |
| 0.0 | 1.1 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 0.6 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.3 | GO:0016121 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.0 | 0.2 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.3 | GO:0030431 | sleep(GO:0030431) |
| 0.0 | 0.8 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.8 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.0 | 0.5 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.2 | GO:0010460 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) |
| 0.0 | 0.9 | GO:0050708 | regulation of protein secretion(GO:0050708) |
| 0.0 | 0.4 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 0.1 | GO:1902767 | farnesyl diphosphate biosynthetic process, mevalonate pathway(GO:0010142) isoprenoid biosynthetic process via mevalonate(GO:1902767) |
| 0.0 | 0.7 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.6 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.3 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.0 | 0.3 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 0.6 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.2 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.6 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.7 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.2 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 1.2 | GO:0031023 | microtubule organizing center organization(GO:0031023) |
| 0.0 | 0.2 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 0.1 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.4 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.4 | GO:0061028 | establishment of endothelial barrier(GO:0061028) |
| 0.0 | 0.9 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.2 | 1.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.4 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.9 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.4 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.2 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 0.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 1.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.3 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.0 | 0.8 | GO:0071010 | U2-type prespliceosome(GO:0071004) prespliceosome(GO:0071010) |
| 0.0 | 0.7 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.2 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.0 | 1.1 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.0 | 1.0 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.4 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 0.4 | GO:0008518 | reduced folate carrier activity(GO:0008518) methotrexate transporter activity(GO:0015350) |
| 0.1 | 0.5 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.1 | 1.8 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 0.4 | GO:0017020 | myosin phosphatase regulator activity(GO:0017020) |
| 0.1 | 1.6 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.9 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 0.6 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.1 | 0.4 | GO:0019202 | amino acid kinase activity(GO:0019202) |
| 0.1 | 0.9 | GO:0005222 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 0.2 | GO:0038046 | enkephalin receptor activity(GO:0038046) |
| 0.1 | 0.6 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.1 | 0.5 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 0.3 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 0.9 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.1 | 0.8 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 0.3 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 0.9 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.9 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.2 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 0.3 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.0 | 1.0 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.6 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.3 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.4 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.4 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.4 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.0 | 0.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 1.1 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.6 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.3 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.2 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.2 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.0 | 0.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.8 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.7 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 0.6 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 1.0 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 1.4 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.6 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 1.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 0.8 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.7 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 0.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.5 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.2 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.3 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |