Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
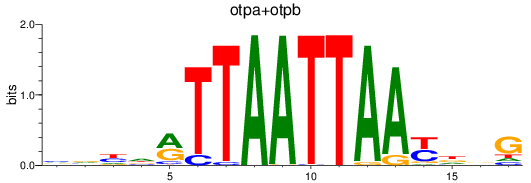
Results for otpa+otpb
Z-value: 0.47
Transcription factors associated with otpa+otpb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
otpa
|
ENSDARG00000014201 | orthopedia homeobox a |
|
otpb
|
ENSDARG00000058379 | orthopedia homeobox b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| otpa | dr11_v1_chr21_+_7582036_7582059 | -0.32 | 1.8e-01 | Click! |
| otpb | dr11_v1_chr5_-_51619262_51619321 | 0.10 | 6.9e-01 | Click! |
Activity profile of otpa+otpb motif
Sorted Z-values of otpa+otpb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_54209504 | 1.74 |
ENSDART00000020033
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr12_-_35830625 | 1.56 |
ENSDART00000180028
|
CU459056.1
|
|
| chr1_-_31534089 | 1.50 |
ENSDART00000007770
|
lbx1b
|
ladybird homeobox 1b |
| chr17_-_16422654 | 1.48 |
ENSDART00000150149
|
tdp1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr20_+_98179 | 1.11 |
ENSDART00000022725
|
si:ch1073-155h21.1
|
si:ch1073-155h21.1 |
| chr23_+_36460239 | 1.04 |
ENSDART00000172441
|
lima1a
|
LIM domain and actin binding 1a |
| chr3_-_15999501 | 0.99 |
ENSDART00000160668
|
nme3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr3_-_19368435 | 0.95 |
ENSDART00000132987
|
s1pr5a
|
sphingosine-1-phosphate receptor 5a |
| chr7_+_38811800 | 0.93 |
ENSDART00000052322
|
zgc:110699
|
zgc:110699 |
| chr24_+_19415124 | 0.92 |
ENSDART00000186931
|
sulf1
|
sulfatase 1 |
| chr3_+_6469754 | 0.85 |
ENSDART00000185809
|
NUP85 (1 of many)
|
nucleoporin 85 |
| chr9_+_37152564 | 0.82 |
ENSDART00000189497
|
gli2a
|
GLI family zinc finger 2a |
| chr4_+_57881965 | 0.81 |
ENSDART00000162234
|
si:dkeyp-44b5.4
|
si:dkeyp-44b5.4 |
| chr25_+_22320738 | 0.79 |
ENSDART00000073566
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr20_+_11731039 | 0.78 |
ENSDART00000152215
ENSDART00000152585 |
si:ch211-155o21.3
|
si:ch211-155o21.3 |
| chr23_-_16485190 | 0.78 |
ENSDART00000155038
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr1_+_21731382 | 0.74 |
ENSDART00000054395
|
pax5
|
paired box 5 |
| chr22_-_16154771 | 0.74 |
ENSDART00000009464
|
slc30a7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr7_-_30174882 | 0.72 |
ENSDART00000110409
|
frmd5
|
FERM domain containing 5 |
| chr11_-_10456553 | 0.72 |
ENSDART00000169509
ENSDART00000185574 ENSDART00000188276 |
ect2
|
epithelial cell transforming 2 |
| chr1_-_45616470 | 0.72 |
ENSDART00000150165
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr5_+_67390115 | 0.72 |
ENSDART00000193255
|
ebf2
|
early B cell factor 2 |
| chr9_-_27398369 | 0.72 |
ENSDART00000186499
|
tex30
|
testis expressed 30 |
| chr14_+_989733 | 0.70 |
ENSDART00000161487
ENSDART00000127317 |
si:ch73-308l14.2
|
si:ch73-308l14.2 |
| chr9_+_48761455 | 0.70 |
ENSDART00000139631
|
abcb11a
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11a |
| chr25_-_21031007 | 0.68 |
ENSDART00000138985
|
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr11_+_44617021 | 0.68 |
ENSDART00000158887
|
rbm34
|
RNA binding motif protein 34 |
| chr22_+_508290 | 0.66 |
ENSDART00000135403
|
nuak2
|
NUAK family, SNF1-like kinase, 2 |
| chr11_-_10456387 | 0.65 |
ENSDART00000011087
ENSDART00000081827 |
ect2
|
epithelial cell transforming 2 |
| chr5_+_60590796 | 0.65 |
ENSDART00000159859
|
tmem132e
|
transmembrane protein 132E |
| chr22_-_14247276 | 0.64 |
ENSDART00000033332
|
si:ch211-246m6.5
|
si:ch211-246m6.5 |
| chr6_-_33878665 | 0.63 |
ENSDART00000129916
|
tmem69
|
transmembrane protein 69 |
| chr1_+_52792439 | 0.62 |
ENSDART00000123972
|
smarca5
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 |
| chr11_+_34760628 | 0.62 |
ENSDART00000087216
|
si:dkey-202e22.2
|
si:dkey-202e22.2 |
| chr23_+_1029450 | 0.61 |
ENSDART00000189196
|
si:zfos-905g2.1
|
si:zfos-905g2.1 |
| chr2_-_30668580 | 0.60 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr18_+_14619544 | 0.60 |
ENSDART00000010821
|
utp4
|
UTP4, small subunit processome component |
| chr21_+_25236297 | 0.59 |
ENSDART00000112783
|
tmem45b
|
transmembrane protein 45B |
| chr14_-_26425416 | 0.59 |
ENSDART00000088690
|
lman2
|
lectin, mannose-binding 2 |
| chr1_-_669717 | 0.57 |
ENSDART00000160564
|
cyyr1
|
cysteine/tyrosine-rich 1 |
| chr19_+_43359075 | 0.55 |
ENSDART00000148287
ENSDART00000149856 ENSDART00000188236 ENSDART00000136695 ENSDART00000193859 |
yrk
|
Yes-related kinase |
| chr5_+_41477954 | 0.54 |
ENSDART00000185871
|
pias2
|
protein inhibitor of activated STAT, 2 |
| chr18_+_17725410 | 0.53 |
ENSDART00000090608
|
rspry1
|
ring finger and SPRY domain containing 1 |
| chr25_+_2263857 | 0.52 |
ENSDART00000076439
|
yars2
|
tyrosyl-tRNA synthetase 2, mitochondrial |
| chr24_+_28953089 | 0.51 |
ENSDART00000153761
|
rnpc3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr8_-_33381913 | 0.51 |
ENSDART00000076420
|
lmx1bb
|
LIM homeobox transcription factor 1, beta b |
| chr25_-_27722614 | 0.50 |
ENSDART00000190154
|
zgc:153935
|
zgc:153935 |
| chr6_-_35046735 | 0.50 |
ENSDART00000143649
|
uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr23_-_16484383 | 0.49 |
ENSDART00000187839
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr18_+_41527877 | 0.48 |
ENSDART00000146972
|
selenot1b
|
selenoprotein T, 1b |
| chr5_+_41477526 | 0.48 |
ENSDART00000153567
|
pias2
|
protein inhibitor of activated STAT, 2 |
| chr3_+_13848226 | 0.47 |
ENSDART00000184342
|
ilf3b
|
interleukin enhancer binding factor 3b |
| chr8_+_49065348 | 0.47 |
ENSDART00000032277
|
ddx51
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 51 |
| chr22_-_36530902 | 0.47 |
ENSDART00000056188
|
polr2h
|
info polymerase (RNA) II (DNA directed) polypeptide H |
| chr20_-_45812144 | 0.47 |
ENSDART00000147897
ENSDART00000147637 |
fermt1
|
fermitin family member 1 |
| chr20_+_25586099 | 0.45 |
ENSDART00000063122
ENSDART00000134047 |
cyp2p10
|
cytochrome P450, family 2, subfamily P, polypeptide 10 |
| chr3_+_26342768 | 0.45 |
ENSDART00000163832
|
si:ch211-156b7.4
|
si:ch211-156b7.4 |
| chr8_-_33381430 | 0.45 |
ENSDART00000190019
|
lmx1bb
|
LIM homeobox transcription factor 1, beta b |
| chr1_+_45663727 | 0.44 |
ENSDART00000038574
ENSDART00000141144 ENSDART00000149565 |
trappc5
|
trafficking protein particle complex 5 |
| chr14_-_1355544 | 0.44 |
ENSDART00000060417
|
cetn4
|
centrin 4 |
| chr17_+_41992054 | 0.42 |
ENSDART00000182878
ENSDART00000111537 |
kiz
|
kizuna centrosomal protein |
| chr24_-_10006158 | 0.42 |
ENSDART00000106244
|
zgc:171750
|
zgc:171750 |
| chr17_+_24821627 | 0.41 |
ENSDART00000112389
|
wdr43
|
WD repeat domain 43 |
| chr22_-_10156581 | 0.40 |
ENSDART00000168304
|
rbck1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr9_+_23895711 | 0.40 |
ENSDART00000034686
|
cops8
|
COP9 signalosome subunit 8 |
| chr5_-_12272689 | 0.39 |
ENSDART00000099810
|
nos1
|
nitric oxide synthase 1 (neuronal) |
| chr18_+_18439332 | 0.38 |
ENSDART00000043843
|
siah1
|
siah E3 ubiquitin protein ligase 1 |
| chr10_-_11353469 | 0.38 |
ENSDART00000191847
|
lin54
|
lin-54 DREAM MuvB core complex component |
| chr25_-_28674739 | 0.37 |
ENSDART00000067073
|
lrrc10
|
leucine rich repeat containing 10 |
| chr23_+_11285662 | 0.37 |
ENSDART00000111028
|
chl1a
|
cell adhesion molecule L1-like a |
| chr7_-_34448076 | 0.37 |
ENSDART00000170935
|
nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr6_+_25257728 | 0.36 |
ENSDART00000162581
|
kyat3
|
kynurenine aminotransferase 3 |
| chr11_+_17984354 | 0.36 |
ENSDART00000179986
|
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr21_+_27513859 | 0.36 |
ENSDART00000065420
|
pacs1a
|
phosphofurin acidic cluster sorting protein 1a |
| chr4_+_13449775 | 0.35 |
ENSDART00000172552
|
dyrk2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| chr16_+_25068576 | 0.35 |
ENSDART00000125838
|
im:7147486
|
im:7147486 |
| chr10_+_1681518 | 0.35 |
ENSDART00000018532
|
triap1
|
TP53 regulated inhibitor of apoptosis 1 |
| chr17_+_22577472 | 0.34 |
ENSDART00000045099
|
yipf4
|
Yip1 domain family, member 4 |
| chr6_+_12527725 | 0.33 |
ENSDART00000149328
|
stk24b
|
serine/threonine kinase 24b (STE20 homolog, yeast) |
| chr6_+_25261297 | 0.33 |
ENSDART00000162824
ENSDART00000163490 ENSDART00000157790 ENSDART00000160978 ENSDART00000161545 ENSDART00000159978 |
kyat3
|
kynurenine aminotransferase 3 |
| chr12_+_20587179 | 0.33 |
ENSDART00000170127
|
arsg
|
arylsulfatase G |
| chr1_+_47499888 | 0.33 |
ENSDART00000027624
|
stn1
|
STN1, CST complex subunit |
| chr14_+_44794936 | 0.32 |
ENSDART00000128881
|
zgc:195212
|
zgc:195212 |
| chr11_-_1509773 | 0.31 |
ENSDART00000050762
|
phactr3b
|
phosphatase and actin regulator 3b |
| chr7_+_21272833 | 0.31 |
ENSDART00000052942
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr17_-_21418340 | 0.30 |
ENSDART00000007021
|
atp6v1ba
|
ATPase, H+ transporting, lysosomal, V1 subunit B, member a |
| chr18_-_17529763 | 0.30 |
ENSDART00000191308
|
bbs2
|
Bardet-Biedl syndrome 2 |
| chr3_+_3681116 | 0.30 |
ENSDART00000109618
|
art4
|
ADP-ribosyltransferase 4 (Dombrock blood group) |
| chr6_-_40029423 | 0.30 |
ENSDART00000103230
|
pfkfb4b
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4b |
| chr16_+_28994709 | 0.30 |
ENSDART00000088023
|
gon4l
|
gon-4-like (C. elegans) |
| chr19_+_2631565 | 0.29 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr3_+_26288981 | 0.29 |
ENSDART00000163500
|
rhot1a
|
ras homolog family member T1a |
| chr13_-_12602920 | 0.29 |
ENSDART00000102311
|
lrit3b
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 3b |
| chr17_-_4245902 | 0.29 |
ENSDART00000151851
|
gdf3
|
growth differentiation factor 3 |
| chr2_-_17393216 | 0.28 |
ENSDART00000123137
|
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr7_+_51774502 | 0.27 |
ENSDART00000007767
|
got2b
|
glutamic-oxaloacetic transaminase 2b, mitochondrial |
| chr20_-_9095105 | 0.27 |
ENSDART00000140792
|
oma1
|
OMA1 zinc metallopeptidase |
| chr1_-_51038885 | 0.27 |
ENSDART00000035150
|
spast
|
spastin |
| chr22_-_9183944 | 0.26 |
ENSDART00000188599
|
si:ch211-213a13.5
|
si:ch211-213a13.5 |
| chr24_+_23202545 | 0.26 |
ENSDART00000143862
|
pex2
|
peroxisomal biogenesis factor 2 |
| chr4_-_133492 | 0.25 |
ENSDART00000171845
|
gpr19
|
G protein-coupled receptor 19 |
| chr21_-_1644414 | 0.24 |
ENSDART00000105736
ENSDART00000124904 |
zgc:152948
|
zgc:152948 |
| chr14_+_1170968 | 0.23 |
ENSDART00000125203
ENSDART00000193575 |
hopx
|
HOP homeobox |
| chr23_+_12406722 | 0.23 |
ENSDART00000145008
|
pigt
|
phosphatidylinositol glycan anchor biosynthesis, class T |
| chr11_+_17984167 | 0.22 |
ENSDART00000020283
ENSDART00000188329 |
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr6_+_14980761 | 0.22 |
ENSDART00000087782
|
mrps9
|
mitochondrial ribosomal protein S9 |
| chr16_+_23303859 | 0.22 |
ENSDART00000006093
|
slc50a1
|
solute carrier family 50 (sugar efflux transporter), member 1 |
| chr14_-_470505 | 0.22 |
ENSDART00000067147
|
ANKRD50
|
ankyrin repeat domain 50 |
| chr4_+_19700308 | 0.21 |
ENSDART00000027919
|
pax4
|
paired box 4 |
| chr12_-_31794908 | 0.21 |
ENSDART00000105583
ENSDART00000153449 |
nat9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr8_-_13046089 | 0.20 |
ENSDART00000137784
|
si:dkey-208b23.5
|
si:dkey-208b23.5 |
| chr5_-_11809710 | 0.19 |
ENSDART00000186998
ENSDART00000181363 ENSDART00000180681 |
nf2a
|
neurofibromin 2a (merlin) |
| chr22_+_5532003 | 0.18 |
ENSDART00000106174
|
si:ch73-256j6.2
|
si:ch73-256j6.2 |
| chr9_-_32177117 | 0.18 |
ENSDART00000078568
|
sf3b1
|
splicing factor 3b, subunit 1 |
| chr14_-_1200854 | 0.18 |
ENSDART00000106672
|
arl9
|
ADP-ribosylation factor-like 9 |
| chr18_+_7456597 | 0.18 |
ENSDART00000142270
|
terb1
|
telomere repeat binding bouquet formation protein 1 |
| chr2_-_17392799 | 0.17 |
ENSDART00000136470
ENSDART00000141188 |
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr22_+_2751887 | 0.16 |
ENSDART00000133652
|
si:dkey-20i20.11
|
si:dkey-20i20.11 |
| chr18_+_33272861 | 0.15 |
ENSDART00000131796
|
v2ra20
|
vomeronasal 2 receptor, a20 |
| chr17_-_25831569 | 0.15 |
ENSDART00000148743
|
hhat
|
hedgehog acyltransferase |
| chr14_-_33481428 | 0.15 |
ENSDART00000147059
ENSDART00000140001 ENSDART00000124242 ENSDART00000164836 ENSDART00000190104 ENSDART00000186833 ENSDART00000180873 |
lamp2
|
lysosomal-associated membrane protein 2 |
| chr17_+_20589553 | 0.15 |
ENSDART00000154447
|
si:ch73-288o11.4
|
si:ch73-288o11.4 |
| chr5_+_33498253 | 0.14 |
ENSDART00000140993
|
ms4a17c.2
|
membrane-spanning 4-domains, subfamily A, member 17c.2 |
| chr4_-_67980261 | 0.12 |
ENSDART00000182305
|
si:ch211-223k15.1
|
si:ch211-223k15.1 |
| chr5_-_38451082 | 0.12 |
ENSDART00000136428
|
chrne
|
cholinergic receptor, nicotinic, epsilon |
| chr12_+_36428052 | 0.12 |
ENSDART00000131300
|
unk
|
unkempt family zinc finger |
| chr9_-_3934963 | 0.11 |
ENSDART00000062336
|
ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr25_+_21833287 | 0.11 |
ENSDART00000187606
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr16_+_23796612 | 0.11 |
ENSDART00000131698
|
rab13
|
RAB13, member RAS oncogene family |
| chr25_-_13490744 | 0.11 |
ENSDART00000056721
|
ldhd
|
lactate dehydrogenase D |
| chr15_+_37965953 | 0.10 |
ENSDART00000154192
|
si:dkey-238d18.10
|
si:dkey-238d18.10 |
| chr9_-_25055722 | 0.10 |
ENSDART00000137131
|
dnah7
|
dynein, axonemal, heavy chain 7 |
| chr8_-_53044300 | 0.09 |
ENSDART00000191653
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr15_-_21014270 | 0.09 |
ENSDART00000154019
|
si:ch211-212c13.10
|
si:ch211-212c13.10 |
| chr24_-_16980337 | 0.09 |
ENSDART00000183812
|
klhl15
|
kelch-like family member 15 |
| chr4_-_9891874 | 0.09 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr4_+_59845617 | 0.09 |
ENSDART00000167626
ENSDART00000123157 |
si:dkey-196n19.2
|
si:dkey-196n19.2 |
| chr1_+_51039558 | 0.08 |
ENSDART00000024743
|
dpy30
|
dpy-30 histone methyltransferase complex regulatory subunit |
| chr7_+_20512419 | 0.08 |
ENSDART00000173907
|
si:dkey-19b23.14
|
si:dkey-19b23.14 |
| chr2_-_37140423 | 0.08 |
ENSDART00000144220
|
tspan37
|
tetraspanin 37 |
| chr7_-_57509447 | 0.08 |
ENSDART00000051973
ENSDART00000147036 |
sirt3
|
sirtuin 3 |
| chr20_-_29864390 | 0.08 |
ENSDART00000161834
ENSDART00000132278 |
rnf144ab
|
ring finger protein 144ab |
| chr9_+_24065855 | 0.06 |
ENSDART00000161468
ENSDART00000171577 ENSDART00000172743 ENSDART00000159324 ENSDART00000079689 ENSDART00000023196 ENSDART00000101577 |
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr2_+_37140448 | 0.06 |
ENSDART00000045016
ENSDART00000142940 |
pex19
|
peroxisomal biogenesis factor 19 |
| chr18_+_7456888 | 0.06 |
ENSDART00000081468
|
terb1
|
telomere repeat binding bouquet formation protein 1 |
| chr3_+_23029484 | 0.06 |
ENSDART00000187900
|
nags
|
N-acetylglutamate synthase |
| chr22_-_19552796 | 0.04 |
ENSDART00000148088
ENSDART00000105485 |
si:dkey-78l4.14
|
si:dkey-78l4.14 |
| chr19_+_46158078 | 0.04 |
ENSDART00000183933
ENSDART00000164055 |
cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr15_+_5360407 | 0.04 |
ENSDART00000110420
|
or112-1
|
odorant receptor, family A, subfamily 112, member 1 |
| chr6_-_51771634 | 0.02 |
ENSDART00000073847
|
blcap
|
bladder cancer associated protein |
| chr21_+_38634296 | 0.02 |
ENSDART00000114467
|
AL935199.1
|
|
| chr20_+_23408970 | 0.01 |
ENSDART00000144261
|
fryl
|
furry homolog, like |
| chr18_-_17485419 | 0.00 |
ENSDART00000018764
|
foxl1
|
forkhead box L1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of otpa+otpb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.2 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.2 | 0.8 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.2 | 0.7 | GO:0015722 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.2 | 0.8 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.1 | 0.4 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.1 | 0.4 | GO:0098924 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) retrograde trans-synaptic signaling by nitric oxide(GO:0098924) |
| 0.1 | 0.4 | GO:1904729 | regulation of intestinal absorption(GO:1904478) regulation of intestinal lipid absorption(GO:1904729) |
| 0.1 | 0.9 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.1 | 1.4 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.1 | 0.6 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.1 | 0.3 | GO:0051228 | protein hexamerization(GO:0034214) mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.1 | 0.7 | GO:0060005 | reflex(GO:0060004) vestibular reflex(GO:0060005) vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.1 | 0.4 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 0.2 | GO:0045141 | telomere localization(GO:0034397) telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic telomere clustering(GO:0045141) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 1.0 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.1 | 0.4 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.1 | 0.3 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.1 | 0.2 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.1 | 0.6 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.1 | 1.0 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.1 | 0.3 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.1 | 0.7 | GO:0090309 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.3 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.5 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.2 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 1.0 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.6 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.3 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.3 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 0.4 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.0 | 0.3 | GO:0018120 | peptidyl-arginine ADP-ribosylation(GO:0018120) |
| 0.0 | 0.1 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.7 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.6 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.5 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.6 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.0 | 0.6 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.7 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.1 | GO:0045046 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.2 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.3 | GO:0016233 | telomere capping(GO:0016233) |
| 0.0 | 0.6 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.3 | GO:0045454 | cell redox homeostasis(GO:0045454) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.1 | 0.3 | GO:1990879 | CST complex(GO:1990879) |
| 0.1 | 0.6 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.2 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.0 | 0.4 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.5 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.3 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.7 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.6 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.6 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.3 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.2 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.2 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.3 | 0.8 | GO:0016713 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.2 | 0.7 | GO:0015432 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.1 | 0.5 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.1 | 1.0 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 0.5 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.1 | 1.0 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.3 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.1 | 0.3 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 0.4 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.1 | 0.1 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.1 | 1.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 0.4 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 0.5 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.5 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.6 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 1.7 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 1.0 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.3 | GO:0098847 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.6 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.3 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.5 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.1 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 0.0 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.6 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 1.7 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.4 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.1 | 0.8 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 0.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.5 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.5 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.6 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 1.3 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.4 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.1 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.4 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |