Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
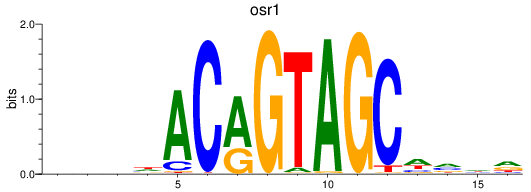
Results for osr1
Z-value: 0.97
Transcription factors associated with osr1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
osr1
|
ENSDARG00000014091 | odd-skipped related transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| osr1 | dr11_v1_chr13_+_32148338_32148338 | -0.31 | 1.9e-01 | Click! |
Activity profile of osr1 motif
Sorted Z-values of osr1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_8040436 | 3.87 |
ENSDART00000113033
|
si:ch211-240l19.6
|
si:ch211-240l19.6 |
| chr11_-_29996344 | 3.65 |
ENSDART00000003712
ENSDART00000126110 |
ace2
|
angiotensin I converting enzyme 2 |
| chr20_+_30490682 | 2.59 |
ENSDART00000184871
|
myt1la
|
myelin transcription factor 1-like, a |
| chr18_+_907266 | 2.50 |
ENSDART00000171729
|
pkma
|
pyruvate kinase M1/2a |
| chr9_-_22069364 | 2.50 |
ENSDART00000101938
|
crygm2b
|
crystallin, gamma M2b |
| chr19_-_1023051 | 2.44 |
ENSDART00000158429
|
tmem42b
|
transmembrane protein 42b |
| chr14_+_35901249 | 2.40 |
ENSDART00000105604
|
zgc:77938
|
zgc:77938 |
| chr13_-_992458 | 2.35 |
ENSDART00000114655
|
BFSP1
|
beaded filament structural protein 1 |
| chr20_-_29418620 | 2.18 |
ENSDART00000172634
|
ryr3
|
ryanodine receptor 3 |
| chr24_+_40529346 | 2.16 |
ENSDART00000168548
|
CU929259.1
|
|
| chr2_+_39021282 | 2.16 |
ENSDART00000056577
|
RBP1 (1 of many)
|
si:ch211-119o8.7 |
| chr5_-_55981288 | 2.13 |
ENSDART00000146616
|
si:dkey-189h5.6
|
si:dkey-189h5.6 |
| chr17_+_31185276 | 2.12 |
ENSDART00000062887
|
disp2
|
dispatched homolog 2 (Drosophila) |
| chr9_-_22023061 | 2.11 |
ENSDART00000101952
|
crygm2c
|
crystallin, gamma M2c |
| chr3_+_19460991 | 2.07 |
ENSDART00000169124
|
pde6ga
|
phosphodiesterase 6G, cGMP-specific, rod, gamma, paralog a |
| chr2_-_11512819 | 2.01 |
ENSDART00000142013
|
penka
|
proenkephalin a |
| chr3_-_61181018 | 1.99 |
ENSDART00000187970
|
pvalb4
|
parvalbumin 4 |
| chr5_+_30682737 | 1.98 |
ENSDART00000172886
|
pdzd3b
|
PDZ domain containing 3b |
| chr3_-_30123113 | 1.88 |
ENSDART00000153562
|
ppfia3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
| chr23_-_30785382 | 1.88 |
ENSDART00000136156
ENSDART00000131285 |
myt1a
|
myelin transcription factor 1a |
| chr1_-_21344478 | 1.87 |
ENSDART00000077805
|
gria2a
|
glutamate receptor, ionotropic, AMPA 2a |
| chr10_+_2587234 | 1.84 |
ENSDART00000126937
|
wu:fb59d01
|
wu:fb59d01 |
| chr15_-_462783 | 1.77 |
ENSDART00000161049
|
CABZ01033205.1
|
Danio rerio uncharacterized LOC100002960 (LOC100002960), mRNA. |
| chr20_+_1412193 | 1.75 |
ENSDART00000064419
|
leg1.1
|
liver-enriched gene 1, tandem duplicate 1 |
| chr15_-_576135 | 1.75 |
ENSDART00000124170
|
cbln20
|
cerebellin 20 |
| chr14_+_4151379 | 1.74 |
ENSDART00000160431
|
dhrs13l1
|
dehydrogenase/reductase (SDR family) member 13 like 1 |
| chr1_-_8928841 | 1.70 |
ENSDART00000103652
|
grin2ab
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2A, b |
| chr14_+_49135264 | 1.67 |
ENSDART00000084119
|
si:ch1073-44g3.1
|
si:ch1073-44g3.1 |
| chr8_-_53896982 | 1.63 |
ENSDART00000169206
|
MIB2
|
mindbomb E3 ubiquitin protein ligase 2 |
| chr11_-_37548711 | 1.62 |
ENSDART00000127184
|
bsnb
|
bassoon (presynaptic cytomatrix protein) b |
| chr1_-_34750169 | 1.60 |
ENSDART00000149380
|
si:dkey-151m22.8
|
si:dkey-151m22.8 |
| chr16_-_27749172 | 1.58 |
ENSDART00000145198
|
steap4
|
STEAP family member 4 |
| chr10_-_2526526 | 1.57 |
ENSDART00000191726
ENSDART00000192767 |
CU856539.1
|
|
| chr12_+_31713239 | 1.55 |
ENSDART00000122379
|
habp2
|
hyaluronan binding protein 2 |
| chr8_-_33114202 | 1.53 |
ENSDART00000098840
|
ralgps1
|
Ral GEF with PH domain and SH3 binding motif 1 |
| chr4_-_67969695 | 1.50 |
ENSDART00000190016
|
si:ch211-223k15.1
|
si:ch211-223k15.1 |
| chr19_-_9503473 | 1.50 |
ENSDART00000091615
|
iffo1a
|
intermediate filament family orphan 1a |
| chr15_+_3219134 | 1.48 |
ENSDART00000113532
|
LO017656.1
|
|
| chr13_-_33022372 | 1.46 |
ENSDART00000147165
|
rbm25a
|
RNA binding motif protein 25a |
| chr25_-_19395476 | 1.44 |
ENSDART00000182622
|
map1ab
|
microtubule-associated protein 1Ab |
| chr21_+_10021823 | 1.43 |
ENSDART00000163995
|
herc7
|
hect domain and RLD 7 |
| chr3_+_23248542 | 1.38 |
ENSDART00000185765
ENSDART00000192332 |
ppp1r9ba
|
protein phosphatase 1, regulatory subunit 9Ba |
| chr2_+_20539402 | 1.37 |
ENSDART00000129585
|
si:ch73-14h1.2
|
si:ch73-14h1.2 |
| chr22_-_28871787 | 1.34 |
ENSDART00000104828
|
gtpbp2b
|
GTP binding protein 2b |
| chr1_+_44826593 | 1.34 |
ENSDART00000162200
|
STX3
|
zgc:165520 |
| chr7_+_29954709 | 1.34 |
ENSDART00000173904
|
tpma
|
alpha-tropomyosin |
| chr2_+_50626476 | 1.33 |
ENSDART00000018150
|
neurod6b
|
neuronal differentiation 6b |
| chr2_+_3986083 | 1.32 |
ENSDART00000188979
|
mkxb
|
mohawk homeobox b |
| chr20_-_5291012 | 1.31 |
ENSDART00000122892
|
cyp46a1.3
|
cytochrome P450, family 46, subfamily A, polypeptide 1, tandem duplicate 3 |
| chr13_-_40499296 | 1.27 |
ENSDART00000158338
|
CNNM1
|
Danio rerio cyclin and CBS domain divalent metal cation transport mediator 1 (cnnm1), mRNA. |
| chr2_-_9646857 | 1.26 |
ENSDART00000056901
|
zgc:153615
|
zgc:153615 |
| chr13_-_11536951 | 1.25 |
ENSDART00000018155
|
adss
|
adenylosuccinate synthase |
| chr23_+_43809484 | 1.24 |
ENSDART00000149534
|
si:ch211-149b19.3
|
si:ch211-149b19.3 |
| chr1_-_58064738 | 1.23 |
ENSDART00000073778
|
caspb
|
caspase b |
| chr12_-_45533034 | 1.18 |
ENSDART00000113004
|
CABZ01068356.1
|
|
| chr1_+_10771457 | 1.18 |
ENSDART00000027050
ENSDART00000137185 |
cnga3b
|
cyclic nucleotide gated channel alpha 3b |
| chr18_-_43880020 | 1.17 |
ENSDART00000185638
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr10_-_19497914 | 1.17 |
ENSDART00000132084
|
si:ch211-127i16.2
|
si:ch211-127i16.2 |
| chr17_-_26537928 | 1.15 |
ENSDART00000155692
ENSDART00000122366 |
ccser2a
|
coiled-coil serine-rich protein 2a |
| chr13_-_3351708 | 1.12 |
ENSDART00000042875
|
si:ch73-193i2.2
|
si:ch73-193i2.2 |
| chr10_+_21559605 | 1.12 |
ENSDART00000123648
ENSDART00000108584 |
pcdh1a3
pcdh1a3
|
protocadherin 1 alpha 3 protocadherin 1 alpha 3 |
| chr18_+_1703984 | 1.11 |
ENSDART00000114010
|
slitrk3a
|
SLIT and NTRK-like family, member 3a |
| chr8_-_22639794 | 1.10 |
ENSDART00000188029
|
iqsec2a
|
IQ motif and Sec7 domain 2a |
| chr25_+_35212919 | 1.09 |
ENSDART00000180127
|
ano5a
|
anoctamin 5a |
| chr20_+_43113465 | 1.08 |
ENSDART00000004842
|
dusp23a
|
dual specificity phosphatase 23a |
| chr22_-_11626014 | 1.08 |
ENSDART00000063133
ENSDART00000160085 |
gcga
|
glucagon a |
| chr12_-_7824291 | 1.08 |
ENSDART00000148673
ENSDART00000149453 |
ank3b
|
ankyrin 3b |
| chr5_-_43682930 | 1.07 |
ENSDART00000075017
|
si:dkey-40c11.1
|
si:dkey-40c11.1 |
| chr4_+_3287819 | 1.07 |
ENSDART00000168633
|
CABZ01085700.1
|
|
| chr21_-_40083432 | 1.05 |
ENSDART00000141160
ENSDART00000191195 |
slc13a5a
|
info solute carrier family 13 (sodium-dependent citrate transporter), member 5a |
| chr18_+_21408794 | 1.05 |
ENSDART00000140161
|
necab2
|
N-terminal EF-hand calcium binding protein 2 |
| chr13_+_30696286 | 1.04 |
ENSDART00000192411
|
cxcl18a.1
|
chemokine (C-X-C motif) ligand 18a, duplicate 1 |
| chr2_+_37897079 | 1.04 |
ENSDART00000141784
|
tep1
|
telomerase-associated protein 1 |
| chr17_-_22021213 | 1.02 |
ENSDART00000078843
|
zmp:0000001102
|
zmp:0000001102 |
| chr7_-_4036875 | 1.02 |
ENSDART00000165021
|
ndrg2
|
NDRG family member 2 |
| chr15_-_12011202 | 1.02 |
ENSDART00000160427
ENSDART00000168715 |
si:dkey-202l22.6
|
si:dkey-202l22.6 |
| chr14_+_33955807 | 1.01 |
ENSDART00000173316
|
si:ch73-22o18.1
|
si:ch73-22o18.1 |
| chr7_-_11605185 | 1.01 |
ENSDART00000169291
ENSDART00000113904 |
stard5
|
StAR-related lipid transfer (START) domain containing 5 |
| chr15_+_22390076 | 0.99 |
ENSDART00000183764
|
oafa
|
OAF homolog a (Drosophila) |
| chr21_-_13662237 | 0.98 |
ENSDART00000091647
ENSDART00000151547 |
pnpla7a
|
patatin-like phospholipase domain containing 7a |
| chr20_+_46560258 | 0.97 |
ENSDART00000122115
|
si:ch211-153j24.3
|
si:ch211-153j24.3 |
| chr22_-_19102256 | 0.97 |
ENSDART00000171866
ENSDART00000166295 |
polrmt
|
polymerase (RNA) mitochondrial (DNA directed) |
| chr10_+_29137482 | 0.96 |
ENSDART00000178280
|
picalma
|
phosphatidylinositol binding clathrin assembly protein a |
| chr7_-_71758613 | 0.95 |
ENSDART00000166724
|
myom1b
|
myomesin 1b |
| chr21_+_21195487 | 0.94 |
ENSDART00000181746
ENSDART00000184832 |
rictorb
|
RPTOR independent companion of MTOR, complex 2b |
| chr11_+_21053488 | 0.93 |
ENSDART00000189860
|
zgc:113307
|
zgc:113307 |
| chr2_+_35421285 | 0.93 |
ENSDART00000143787
|
tnr
|
tenascin R (restrictin, janusin) |
| chr7_+_29955368 | 0.93 |
ENSDART00000173686
|
tpma
|
alpha-tropomyosin |
| chr3_-_4303262 | 0.92 |
ENSDART00000112819
|
si:dkey-73p2.2
|
si:dkey-73p2.2 |
| chr11_+_25735478 | 0.92 |
ENSDART00000103566
|
si:dkey-183j2.10
|
si:dkey-183j2.10 |
| chr2_-_57076687 | 0.91 |
ENSDART00000161523
|
slc25a42
|
solute carrier family 25, member 42 |
| chr10_-_13239367 | 0.91 |
ENSDART00000001253
|
si:busm1-57f23.1
|
si:busm1-57f23.1 |
| chr19_-_6134802 | 0.91 |
ENSDART00000140051
|
cica
|
capicua transcriptional repressor a |
| chr1_-_52201266 | 0.91 |
ENSDART00000143805
ENSDART00000023757 |
rab3da
|
RAB3D, member RAS oncogene family, a |
| chr14_-_33872092 | 0.91 |
ENSDART00000111903
|
si:ch73-335m24.2
|
si:ch73-335m24.2 |
| chr16_+_35344031 | 0.90 |
ENSDART00000167140
|
si:dkey-34d22.1
|
si:dkey-34d22.1 |
| chr3_-_26115558 | 0.89 |
ENSDART00000078671
ENSDART00000124762 |
hsp70.1
|
heat shock cognate 70-kd protein, tandem duplicate 1 |
| chr3_+_17744339 | 0.89 |
ENSDART00000132622
|
znf385c
|
zinc finger protein 385C |
| chr9_-_18568927 | 0.87 |
ENSDART00000084668
|
enox1
|
ecto-NOX disulfide-thiol exchanger 1 |
| chr5_-_16996482 | 0.86 |
ENSDART00000144501
|
galnt9
|
polypeptide N-acetylgalactosaminyltransferase 9 |
| chr10_-_5814101 | 0.85 |
ENSDART00000161903
|
il6st
|
interleukin 6 signal transducer |
| chr8_+_49778756 | 0.85 |
ENSDART00000083790
|
ntrk2a
|
neurotrophic tyrosine kinase, receptor, type 2a |
| chr9_-_39547907 | 0.83 |
ENSDART00000163635
|
erbb4b
|
erb-b2 receptor tyrosine kinase 4b |
| chr19_+_14921000 | 0.83 |
ENSDART00000144052
|
oprd1a
|
opioid receptor, delta 1a |
| chr20_-_26866111 | 0.83 |
ENSDART00000077767
|
mylk4b
|
myosin light chain kinase family, member 4b |
| chr14_-_2203124 | 0.83 |
ENSDART00000124171
|
si:dkeyp-115d7.2
|
si:dkeyp-115d7.2 |
| chr25_+_7670683 | 0.82 |
ENSDART00000040275
|
kcnj11l
|
potassium inwardly-rectifying channel, subfamily J, member 11, like |
| chr5_-_16274058 | 0.82 |
ENSDART00000090684
|
kremen1
|
kringle containing transmembrane protein 1 |
| chr20_+_53522059 | 0.82 |
ENSDART00000147570
|
pak6b
|
p21 protein (Cdc42/Rac)-activated kinase 6b |
| chr1_-_7912932 | 0.81 |
ENSDART00000147383
|
mmd2b
|
monocyte to macrophage differentiation-associated 2b |
| chr25_-_19395156 | 0.81 |
ENSDART00000155335
|
map1ab
|
microtubule-associated protein 1Ab |
| chr13_+_25396896 | 0.80 |
ENSDART00000041257
|
gsto2
|
glutathione S-transferase omega 2 |
| chr9_-_5263947 | 0.80 |
ENSDART00000088342
|
cytip
|
cytohesin 1 interacting protein |
| chr9_-_34269066 | 0.79 |
ENSDART00000059955
|
ildr1b
|
immunoglobulin-like domain containing receptor 1b |
| chr16_-_26537103 | 0.79 |
ENSDART00000134908
ENSDART00000008152 |
sgk2b
|
serum/glucocorticoid regulated kinase 2b |
| chr7_-_42206720 | 0.79 |
ENSDART00000110907
|
itfg1
|
integrin alpha FG-GAP repeat containing 1 |
| chr9_-_4856767 | 0.79 |
ENSDART00000016814
ENSDART00000138015 |
fmnl2a
|
formin-like 2a |
| chr11_-_33960318 | 0.78 |
ENSDART00000087597
|
col6a2
|
collagen, type VI, alpha 2 |
| chr3_+_61896514 | 0.77 |
ENSDART00000113524
ENSDART00000158997 |
fam20ca
|
family with sequence similarity 20, member Ca |
| chr1_+_9886991 | 0.77 |
ENSDART00000135702
|
rgs11
|
regulator of G protein signaling 11 |
| chr25_+_37435720 | 0.76 |
ENSDART00000164390
|
chmp1a
|
charged multivesicular body protein 1A |
| chr19_-_35361556 | 0.76 |
ENSDART00000012167
|
ndufs5
|
NADH dehydrogenase (ubiquinone) Fe-S protein 5 |
| chr7_+_20656942 | 0.74 |
ENSDART00000100898
|
tnfsf12
|
TNF superfamily member 12 |
| chr7_+_22767678 | 0.73 |
ENSDART00000137203
|
ponzr6
|
plac8 onzin related protein 6 |
| chr8_-_4100365 | 0.73 |
ENSDART00000142846
|
cux2b
|
cut-like homeobox 2b |
| chr7_-_17771166 | 0.72 |
ENSDART00000173620
|
si:dkey-106g10.7
|
si:dkey-106g10.7 |
| chr23_-_42752550 | 0.71 |
ENSDART00000187059
|
si:ch73-217n20.1
|
si:ch73-217n20.1 |
| chr2_-_15318786 | 0.71 |
ENSDART00000135851
|
tecrl2b
|
trans-2,3-enoyl-CoA reductase-like 2b |
| chr10_-_25860102 | 0.71 |
ENSDART00000080789
|
trpc4a
|
transient receptor potential cation channel, subfamily C, member 4a |
| chr6_+_52947186 | 0.71 |
ENSDART00000155831
|
uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr21_-_37951819 | 0.71 |
ENSDART00000139549
|
si:dkey-38k9.5
|
si:dkey-38k9.5 |
| chr13_+_25397098 | 0.70 |
ENSDART00000132953
|
gsto2
|
glutathione S-transferase omega 2 |
| chr13_+_50151407 | 0.70 |
ENSDART00000031858
|
gpr137ba
|
G protein-coupled receptor 137Ba |
| chr13_-_27038407 | 0.69 |
ENSDART00000146712
|
ccdc85a
|
coiled-coil domain containing 85A |
| chr7_-_57949117 | 0.69 |
ENSDART00000138803
|
ank2b
|
ankyrin 2b, neuronal |
| chr19_-_25519310 | 0.69 |
ENSDART00000089882
|
C1GALT1 (1 of many)
|
si:dkey-202e17.1 |
| chr19_+_10794072 | 0.68 |
ENSDART00000151518
ENSDART00000142614 |
lipea
|
lipase, hormone-sensitive a |
| chr13_+_23132666 | 0.68 |
ENSDART00000164639
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr10_+_29138021 | 0.68 |
ENSDART00000025227
ENSDART00000123033 ENSDART00000034242 |
picalma
|
phosphatidylinositol binding clathrin assembly protein a |
| chr12_-_15620090 | 0.67 |
ENSDART00000038032
|
acbd4
|
acyl-CoA binding domain containing 4 |
| chr4_-_6567355 | 0.67 |
ENSDART00000134820
ENSDART00000142087 |
foxp2
|
forkhead box P2 |
| chr15_-_25392589 | 0.66 |
ENSDART00000124205
|
si:dkey-54n8.4
|
si:dkey-54n8.4 |
| chr19_-_5769553 | 0.66 |
ENSDART00000175003
|
si:ch211-264f5.6
|
si:ch211-264f5.6 |
| chr2_-_14387335 | 0.66 |
ENSDART00000189332
ENSDART00000164786 ENSDART00000188572 |
sgip1b
|
SH3-domain GRB2-like (endophilin) interacting protein 1b |
| chr25_+_34581494 | 0.65 |
ENSDART00000167690
|
trpm1b
|
transient receptor potential cation channel, subfamily M, member 1b |
| chr20_+_16881883 | 0.63 |
ENSDART00000130107
|
nfkbiaa
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha a |
| chr24_-_9960290 | 0.63 |
ENSDART00000143390
ENSDART00000092975 ENSDART00000184953 |
vps41
|
vacuolar protein sorting 41 homolog (S. cerevisiae) |
| chr21_+_39365920 | 0.63 |
ENSDART00000140940
|
cluhb
|
clustered mitochondria (cluA/CLU1) homolog b |
| chr2_-_9748039 | 0.62 |
ENSDART00000134870
|
si:ch1073-170o4.1
|
si:ch1073-170o4.1 |
| chr3_-_10751491 | 0.62 |
ENSDART00000016351
|
zgc:112965
|
zgc:112965 |
| chr5_+_69747417 | 0.62 |
ENSDART00000153717
|
si:ch211-275j6.5
|
si:ch211-275j6.5 |
| chr5_+_69808763 | 0.62 |
ENSDART00000143482
|
fsd1l
|
fibronectin type III and SPRY domain containing 1-like |
| chr15_-_1843831 | 0.62 |
ENSDART00000156718
ENSDART00000154175 |
taf15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr15_-_5016039 | 0.61 |
ENSDART00000156458
|
defbl3
|
defensin, beta-like 3 |
| chr8_+_3496204 | 0.61 |
ENSDART00000085993
|
pxnb
|
paxillin b |
| chr17_-_23631400 | 0.61 |
ENSDART00000079563
|
fas
|
Fas cell surface death receptor |
| chr2_-_49557932 | 0.61 |
ENSDART00000141902
|
si:ch211-209f23.3
|
si:ch211-209f23.3 |
| chr22_+_26804197 | 0.60 |
ENSDART00000135688
|
si:dkey-44g23.2
|
si:dkey-44g23.2 |
| chr10_+_19554604 | 0.59 |
ENSDART00000063806
|
atp6v1b2
|
ATPase H+ transporting V1 subunit B2 |
| chr8_+_30709685 | 0.59 |
ENSDART00000133989
|
upb1
|
ureidopropionase, beta |
| chr1_+_44800298 | 0.58 |
ENSDART00000073712
ENSDART00000187054 |
tmem187
|
transmembrane protein 187 |
| chr23_-_42752387 | 0.57 |
ENSDART00000149781
|
si:ch73-217n20.1
|
si:ch73-217n20.1 |
| chr15_-_4415917 | 0.57 |
ENSDART00000062874
|
atp1b3b
|
ATPase Na+/K+ transporting subunit beta 3b |
| chr7_+_28006096 | 0.57 |
ENSDART00000173843
|
tub
|
tubby bipartite transcription factor |
| chr15_-_38154616 | 0.56 |
ENSDART00000099392
|
irgq2
|
immunity-related GTPase family, q2 |
| chr8_-_36469117 | 0.56 |
ENSDART00000111240
|
mhc2dab
|
major histocompatibility complex class II DAB gene |
| chr2_+_49799470 | 0.55 |
ENSDART00000146325
|
si:ch211-190k17.19
|
si:ch211-190k17.19 |
| chr14_+_17137023 | 0.55 |
ENSDART00000080712
|
slc43a3b
|
solute carrier family 43, member 3b |
| chr15_-_434503 | 0.55 |
ENSDART00000122286
|
CABZ01056629.1
|
|
| chr2_-_51096647 | 0.54 |
ENSDART00000167172
|
THEM6
|
si:ch73-52e5.2 |
| chr5_-_5831037 | 0.54 |
ENSDART00000112856
|
zmp:0000000846
|
zmp:0000000846 |
| chr11_-_41130239 | 0.53 |
ENSDART00000173268
|
dnajc11b
|
DnaJ (Hsp40) homolog, subfamily C, member 11b |
| chr3_-_29899172 | 0.53 |
ENSDART00000140518
ENSDART00000020311 |
rpl27
|
ribosomal protein L27 |
| chr2_+_1001560 | 0.53 |
ENSDART00000134656
|
cacna1eb
|
calcium channel, voltage-dependent, R type, alpha 1E subunit b |
| chr13_+_12456412 | 0.52 |
ENSDART00000057762
|
dapp1
|
dual adaptor of phosphotyrosine and 3-phosphoinositides |
| chr21_+_45627775 | 0.52 |
ENSDART00000185466
|
irf1b
|
interferon regulatory factor 1b |
| chr5_+_6617401 | 0.52 |
ENSDART00000060532
|
zgc:110796
|
zgc:110796 |
| chr3_-_30384353 | 0.50 |
ENSDART00000186832
|
lrrc4ba
|
leucine rich repeat containing 4Ba |
| chr6_-_41229787 | 0.50 |
ENSDART00000065013
|
synpr
|
synaptoporin |
| chr3_+_32129632 | 0.50 |
ENSDART00000174522
|
zgc:109934
|
zgc:109934 |
| chr5_+_25317061 | 0.49 |
ENSDART00000170097
|
trpm6
|
transient receptor potential cation channel, subfamily M, member 6 |
| chr6_-_48087152 | 0.48 |
ENSDART00000180614
|
slc2a1b
|
solute carrier family 2 (facilitated glucose transporter), member 1b |
| chr3_+_23063718 | 0.48 |
ENSDART00000140225
ENSDART00000184431 |
b4galnt2.1
|
beta-1,4-N-acetyl-galactosaminyl transferase 2, tandem duplicate 1 |
| chr7_+_42206847 | 0.48 |
ENSDART00000149250
|
phkb
|
phosphorylase kinase, beta |
| chr7_+_38090515 | 0.47 |
ENSDART00000131387
|
cebpg
|
CCAAT/enhancer binding protein (C/EBP), gamma |
| chr17_+_10094063 | 0.47 |
ENSDART00000168055
|
sec23a
|
Sec23 homolog A, COPII coat complex component |
| chr23_+_38953545 | 0.47 |
ENSDART00000184621
|
ATP9A
|
ATPase phospholipid transporting 9A (putative) |
| chr18_-_16395668 | 0.47 |
ENSDART00000186004
|
mgat4c
|
mgat4 family, member C |
| chr6_+_52947699 | 0.46 |
ENSDART00000180913
|
uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr12_+_2993523 | 0.45 |
ENSDART00000082297
|
lrrc45
|
leucine rich repeat containing 45 |
| chr4_-_13156971 | 0.45 |
ENSDART00000182164
|
grip1
|
glutamate receptor interacting protein 1 |
| chr10_+_10677697 | 0.45 |
ENSDART00000188705
|
fam163b
|
family with sequence similarity 163, member B |
| chr14_-_24391424 | 0.44 |
ENSDART00000113376
ENSDART00000126894 |
fam13b
|
family with sequence similarity 13, member B |
| chr10_+_33744098 | 0.44 |
ENSDART00000147775
|
rxfp2a
|
relaxin/insulin-like family peptide receptor 2a |
| chr14_-_1538600 | 0.44 |
ENSDART00000180925
|
CABZ01109480.1
|
|
| chr3_+_54776475 | 0.43 |
ENSDART00000142710
|
si:ch211-74m13.1
|
si:ch211-74m13.1 |
| chr7_-_55292116 | 0.43 |
ENSDART00000122603
|
rnf166
|
ring finger protein 166 |
| chr21_-_28737320 | 0.43 |
ENSDART00000098696
ENSDART00000124826 ENSDART00000125652 |
nrg2a
|
neuregulin 2a |
| chr21_-_40834413 | 0.42 |
ENSDART00000148513
|
limk1b
|
LIM domain kinase 1b |
| chr24_+_16393302 | 0.42 |
ENSDART00000188670
ENSDART00000081759 ENSDART00000177790 |
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
Network of associatons between targets according to the STRING database.
First level regulatory network of osr1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:0015677 | copper ion import(GO:0015677) |
| 0.3 | 2.1 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.3 | 2.4 | GO:0046292 | formaldehyde metabolic process(GO:0046292) formaldehyde catabolic process(GO:0046294) |
| 0.3 | 0.8 | GO:0099551 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.2 | 0.7 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.2 | 0.6 | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 0.2 | 1.3 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.2 | 0.8 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.2 | 0.8 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.2 | 1.7 | GO:1990402 | embryonic liver development(GO:1990402) |
| 0.2 | 0.9 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.2 | 1.7 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.2 | 2.7 | GO:0048899 | anterior lateral line development(GO:0048899) |
| 0.2 | 1.6 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.2 | 1.1 | GO:0032185 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.2 | 0.6 | GO:0070227 | lymphocyte apoptotic process(GO:0070227) |
| 0.1 | 1.0 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.1 | 1.2 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.7 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.9 | GO:0070572 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.1 | 1.0 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 0.5 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.1 | 1.3 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 0.6 | GO:0019483 | beta-alanine metabolic process(GO:0019482) beta-alanine biosynthetic process(GO:0019483) |
| 0.1 | 0.4 | GO:0016038 | absorption of visible light(GO:0016038) |
| 0.1 | 0.4 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.1 | 1.7 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.1 | 1.7 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.1 | 2.2 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.1 | 0.6 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 0.1 | 0.9 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.3 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 2.0 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 0.6 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.2 | GO:0006210 | thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.1 | 2.3 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 1.2 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.1 | 0.4 | GO:0003222 | ventricular trabecula myocardium morphogenesis(GO:0003222) trabecula formation(GO:0060343) heart trabecula formation(GO:0060347) |
| 0.1 | 3.6 | GO:0060402 | cytosolic calcium ion transport(GO:0060401) calcium ion transport into cytosol(GO:0060402) |
| 0.1 | 0.8 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 0.6 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 0.4 | GO:0046959 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.0 | 0.7 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.3 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.5 | GO:0050688 | regulation of defense response to virus(GO:0050688) |
| 0.0 | 2.0 | GO:0035195 | gene silencing by miRNA(GO:0035195) |
| 0.0 | 0.7 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 0.7 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.4 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.1 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.0 | 2.4 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.8 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.5 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 4.6 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.4 | GO:0099639 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.0 | 0.4 | GO:0030819 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.0 | 0.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.6 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.6 | GO:0006415 | translational termination(GO:0006415) |
| 0.0 | 0.8 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.9 | GO:0007006 | mitochondrial membrane organization(GO:0007006) |
| 0.0 | 0.2 | GO:0048532 | anatomical structure arrangement(GO:0048532) |
| 0.0 | 0.2 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.0 | 1.1 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0046099 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine biosynthetic process(GO:0046099) |
| 0.0 | 0.4 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.8 | GO:0060872 | semicircular canal development(GO:0060872) |
| 0.0 | 0.8 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 1.3 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.6 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.7 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.9 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.1 | GO:0072512 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.0 | 0.6 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.0 | 1.6 | GO:0061515 | myeloid cell development(GO:0061515) |
| 0.0 | 1.9 | GO:0099643 | neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
| 0.0 | 1.4 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 1.5 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 1.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 1.1 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.1 | GO:0034434 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.0 | 0.2 | GO:0051452 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.0 | 1.7 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.7 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.1 | GO:0060753 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.1 | GO:0015862 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.5 | GO:0051262 | protein tetramerization(GO:0051262) |
| 0.0 | 0.8 | GO:0043542 | endothelial cell migration(GO:0043542) |
| 0.0 | 0.6 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.6 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.9 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.2 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.3 | 1.2 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.3 | 2.1 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.2 | 0.9 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.2 | 0.5 | GO:0098556 | cytoplasmic side of rough endoplasmic reticulum membrane(GO:0098556) |
| 0.2 | 1.6 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.2 | 1.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 1.6 | GO:0098894 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.1 | 1.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.6 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.1 | 0.6 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.1 | 0.3 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.1 | 0.6 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.3 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 0.6 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.1 | 0.8 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 2.0 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.5 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.9 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.7 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 1.9 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.6 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 1.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.7 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.7 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.4 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 1.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 1.1 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 2.2 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.7 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 1.7 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.5 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.7 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 1.5 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 2.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0010852 | cyclase inhibitor activity(GO:0010852) guanylate cyclase inhibitor activity(GO:0030251) |
| 0.5 | 1.5 | GO:0045174 | glutathione dehydrogenase (ascorbate) activity(GO:0045174) |
| 0.5 | 2.4 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) S-(hydroxymethyl)glutathione dehydrogenase activity(GO:0051903) |
| 0.4 | 2.2 | GO:0048763 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.4 | 1.7 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.4 | 2.5 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.4 | 1.6 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.3 | 0.9 | GO:0071077 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.3 | 0.8 | GO:0060175 | brain-derived neurotrophic factor binding(GO:0048403) brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.3 | 0.8 | GO:0038046 | enkephalin receptor activity(GO:0038046) |
| 0.2 | 0.9 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.2 | 1.3 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.2 | 1.1 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.2 | 1.8 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.2 | 1.0 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.2 | 1.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.2 | 2.1 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.2 | 2.0 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.1 | 0.4 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.1 | 1.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 1.9 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 0.6 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.1 | 1.2 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 3.6 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.1 | 1.2 | GO:0005223 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 1.6 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.1 | 0.7 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 1.7 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 0.5 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 0.8 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.6 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 0.8 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 4.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.7 | GO:0048018 | receptor activator activity(GO:0030546) receptor agonist activity(GO:0048018) |
| 0.0 | 0.3 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 1.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.4 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.7 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 1.0 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.0 | 0.1 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.0 | 1.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 0.6 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.6 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.7 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.2 | GO:0004960 | thromboxane receptor activity(GO:0004960) |
| 0.0 | 0.1 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.0 | 0.4 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.8 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.6 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.8 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 1.4 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 2.3 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.1 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.0 | 0.2 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 1.9 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 1.1 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.5 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.9 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.9 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.1 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.4 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.2 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.4 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 1.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 0.9 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 0.6 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 0.6 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 0.4 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 1.6 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 1.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 0.6 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 1.2 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 1.0 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.5 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.2 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.0 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.0 | 0.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |