Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
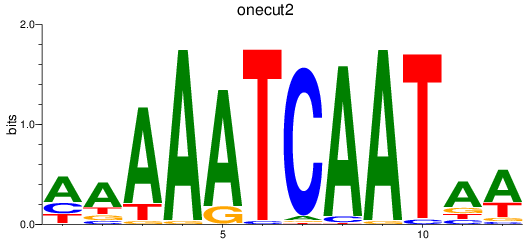
Results for onecut2
Z-value: 0.65
Transcription factors associated with onecut2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
onecut2
|
ENSDARG00000090387 | one cut homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| onecut2 | dr11_v1_chr21_-_1742159_1742159 | 0.52 | 2.3e-02 | Click! |
Activity profile of onecut2 motif
Sorted Z-values of onecut2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_15266093 | 2.77 |
ENSDART00000124676
|
sv2ba
|
synaptic vesicle glycoprotein 2Ba |
| chr11_-_11518469 | 2.36 |
ENSDART00000104254
|
krt15
|
keratin 15 |
| chr12_-_22524388 | 2.26 |
ENSDART00000020942
|
shbg
|
sex hormone-binding globulin |
| chr3_-_28120092 | 2.20 |
ENSDART00000151143
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr17_-_25737452 | 2.15 |
ENSDART00000152021
|
si:ch211-214p16.3
|
si:ch211-214p16.3 |
| chr8_-_23081511 | 2.10 |
ENSDART00000142015
ENSDART00000135764 ENSDART00000147021 |
si:dkey-70p6.1
|
si:dkey-70p6.1 |
| chr5_-_29643930 | 2.08 |
ENSDART00000161250
|
grin1b
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1b |
| chr25_+_7229046 | 1.93 |
ENSDART00000149965
ENSDART00000041820 |
lingo1a
|
leucine rich repeat and Ig domain containing 1a |
| chr2_-_32505091 | 1.92 |
ENSDART00000141884
ENSDART00000056639 |
faim2a
|
Fas apoptotic inhibitory molecule 2a |
| chr6_+_3828560 | 1.87 |
ENSDART00000185273
ENSDART00000179091 |
gad1b
|
glutamate decarboxylase 1b |
| chr1_-_21409877 | 1.83 |
ENSDART00000102782
|
gria2a
|
glutamate receptor, ionotropic, AMPA 2a |
| chr25_-_9805269 | 1.73 |
ENSDART00000192048
|
lrrc4c
|
leucine rich repeat containing 4C |
| chr20_-_40755614 | 1.57 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr18_+_15644559 | 1.43 |
ENSDART00000061794
|
nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr18_+_642889 | 1.40 |
ENSDART00000189007
|
CABZ01078320.1
|
|
| chr16_-_13612650 | 1.34 |
ENSDART00000080372
|
dbpb
|
D site albumin promoter binding protein b |
| chr20_-_40754794 | 1.30 |
ENSDART00000187251
|
cx32.3
|
connexin 32.3 |
| chr14_-_2348917 | 1.27 |
ENSDART00000159004
|
si:ch73-233f7.8
|
si:ch73-233f7.8 |
| chr18_-_37007294 | 1.25 |
ENSDART00000088309
|
map3k10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr7_+_49877148 | 1.25 |
ENSDART00000174182
|
slc1a2a
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2a |
| chr16_-_44127307 | 1.25 |
ENSDART00000104583
ENSDART00000151936 ENSDART00000058685 ENSDART00000190830 |
zfpm2a
|
zinc finger protein, FOG family member 2a |
| chr5_+_27267186 | 1.22 |
ENSDART00000182238
ENSDART00000087857 |
unc5db
|
unc-5 netrin receptor Db |
| chr2_+_7818368 | 1.22 |
ENSDART00000007068
|
kcnmb2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr17_-_45247332 | 1.20 |
ENSDART00000016815
|
ttbk2a
|
tau tubulin kinase 2a |
| chr18_+_50890749 | 1.18 |
ENSDART00000174109
|
si:ch1073-450f2.1
|
si:ch1073-450f2.1 |
| chr4_+_7827261 | 1.14 |
ENSDART00000129568
|
phyh
|
phytanoyl-CoA 2-hydroxylase |
| chr18_-_37007061 | 1.13 |
ENSDART00000136432
|
map3k10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr16_+_5612547 | 1.12 |
ENSDART00000140226
ENSDART00000189352 |
CYTH2
|
si:dkey-283b15.4 |
| chr4_-_8903240 | 1.04 |
ENSDART00000129983
|
mpped1
|
metallophosphoesterase domain containing 1 |
| chr8_-_37263524 | 1.04 |
ENSDART00000061327
|
rh50
|
Rh50-like protein |
| chr8_+_40477264 | 1.02 |
ENSDART00000085559
|
gck
|
glucokinase (hexokinase 4) |
| chr18_+_50276653 | 1.02 |
ENSDART00000192120
|
si:dkey-105e17.1
|
si:dkey-105e17.1 |
| chr21_-_27881752 | 1.01 |
ENSDART00000132583
|
nrxn2a
|
neurexin 2a |
| chr18_+_50276337 | 1.00 |
ENSDART00000140352
|
si:dkey-105e17.1
|
si:dkey-105e17.1 |
| chr5_+_61301525 | 0.99 |
ENSDART00000128773
|
doc2b
|
double C2-like domains, beta |
| chr7_-_41693004 | 0.95 |
ENSDART00000121509
|
malrd1
|
MAM and LDL receptor class A domain containing 1 |
| chr4_-_10599062 | 0.95 |
ENSDART00000048003
|
tspan12
|
tetraspanin 12 |
| chr5_-_65158203 | 0.85 |
ENSDART00000171656
|
sh2d3cb
|
SH2 domain containing 3Cb |
| chr8_-_45279411 | 0.83 |
ENSDART00000175207
|
adamts13
|
ADAM metallopeptidase with thrombospondin type 1 motif, 13 |
| chr11_+_37201483 | 0.82 |
ENSDART00000160930
ENSDART00000173439 ENSDART00000171273 |
zgc:112265
|
zgc:112265 |
| chr1_-_9249943 | 0.82 |
ENSDART00000055011
|
zgc:136472
|
zgc:136472 |
| chr1_-_23157583 | 0.82 |
ENSDART00000144208
|
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr23_-_19434199 | 0.79 |
ENSDART00000132543
|
klhdc8b
|
kelch domain containing 8B |
| chr7_+_20535869 | 0.79 |
ENSDART00000078181
|
zgc:158423
|
zgc:158423 |
| chr19_-_41991104 | 0.76 |
ENSDART00000087055
|
trioa
|
trio Rho guanine nucleotide exchange factor a |
| chr13_-_32648382 | 0.76 |
ENSDART00000138571
ENSDART00000132820 |
bend3
|
BEN domain containing 3 |
| chr3_-_60142530 | 0.76 |
ENSDART00000153247
|
si:ch211-120g10.1
|
si:ch211-120g10.1 |
| chr2_-_6262441 | 0.76 |
ENSDART00000092190
|
arl14
|
ADP-ribosylation factor-like 14 |
| chr5_-_28102698 | 0.74 |
ENSDART00000138140
|
si:ch211-48m9.1
|
si:ch211-48m9.1 |
| chr13_+_36144341 | 0.73 |
ENSDART00000182930
ENSDART00000187327 |
TTC9
|
si:ch211-259k16.3 |
| chr1_+_37391716 | 0.73 |
ENSDART00000191986
|
sparcl1
|
SPARC-like 1 |
| chr3_-_28665291 | 0.72 |
ENSDART00000151670
|
fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr15_-_5467477 | 0.72 |
ENSDART00000123839
|
arrb1
|
arrestin, beta 1 |
| chr3_+_39853788 | 0.72 |
ENSDART00000154869
|
cacna1ha
|
calcium channel, voltage-dependent, T type, alpha 1H subunit a |
| chr4_+_26628822 | 0.71 |
ENSDART00000191030
ENSDART00000186113 ENSDART00000186764 ENSDART00000165158 |
iqsec3a
|
IQ motif and Sec7 domain 3a |
| chr23_+_25832689 | 0.71 |
ENSDART00000138907
|
hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chr4_-_5291256 | 0.71 |
ENSDART00000150864
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr3_-_22228602 | 0.70 |
ENSDART00000017750
|
myl4
|
myosin, light chain 4, alkali; atrial, embryonic |
| chr4_-_8902406 | 0.68 |
ENSDART00000192962
|
mpped1
|
metallophosphoesterase domain containing 1 |
| chr21_-_7687544 | 0.68 |
ENSDART00000134519
|
pde8b
|
phosphodiesterase 8B |
| chr17_-_8169774 | 0.66 |
ENSDART00000091828
|
syne1b
|
spectrin repeat containing, nuclear envelope 1b |
| chr24_+_25692802 | 0.65 |
ENSDART00000190493
|
camk2n2
|
calcium/calmodulin-dependent protein kinase II inhibitor 2 |
| chr17_+_43623598 | 0.63 |
ENSDART00000154138
|
znf365
|
zinc finger protein 365 |
| chr13_-_40141630 | 0.62 |
ENSDART00000181618
ENSDART00000171583 ENSDART00000192780 |
crtac1b
|
cartilage acidic protein 1b |
| chr10_-_15405564 | 0.61 |
ENSDART00000020665
|
sgtb
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr7_+_25036188 | 0.60 |
ENSDART00000163957
ENSDART00000169749 |
sb:cb1058
|
sb:cb1058 |
| chr18_-_46380471 | 0.60 |
ENSDART00000144444
ENSDART00000086772 |
slc19a3b
|
solute carrier family 19 (thiamine transporter), member 3b |
| chr2_-_17044959 | 0.60 |
ENSDART00000090260
|
clcn2a
|
chloride channel, voltage-sensitive 2a |
| chr21_+_4116437 | 0.59 |
ENSDART00000167791
ENSDART00000123759 |
rapgef1b
|
Rap guanine nucleotide exchange factor (GEF) 1b |
| chr5_-_67661102 | 0.59 |
ENSDART00000013605
|
zbtb20
|
zinc finger and BTB domain containing 20 |
| chr15_-_34785594 | 0.58 |
ENSDART00000154256
|
gabbr1a
|
gamma-aminobutyric acid (GABA) B receptor, 1a |
| chr10_-_33156789 | 0.58 |
ENSDART00000192268
ENSDART00000182065 ENSDART00000081170 |
cux1a
|
cut-like homeobox 1a |
| chr14_+_21699414 | 0.57 |
ENSDART00000169942
|
stx3a
|
syntaxin 3A |
| chr8_+_9866351 | 0.57 |
ENSDART00000133985
|
kcnd1
|
potassium voltage-gated channel, Shal-related subfamily, member 1 |
| chr8_+_23745167 | 0.57 |
ENSDART00000149352
|
si:ch211-163l21.11
|
si:ch211-163l21.11 |
| chr25_+_19955598 | 0.56 |
ENSDART00000091547
|
kcna1a
|
potassium voltage-gated channel, shaker-related subfamily, member 1a |
| chr5_+_19309877 | 0.53 |
ENSDART00000190338
|
rusc2
|
RUN and SH3 domain containing 2 |
| chr5_-_63286077 | 0.53 |
ENSDART00000131274
|
tbcelb
|
tubulin folding cofactor E-like b |
| chr19_+_4916233 | 0.53 |
ENSDART00000159512
|
ppp1r1b
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr12_-_25217217 | 0.52 |
ENSDART00000152931
|
kcng3
|
potassium voltage-gated channel, subfamily G, member 3 |
| chr18_+_36769758 | 0.52 |
ENSDART00000180375
ENSDART00000136463 ENSDART00000133487 ENSDART00000130206 |
fosb
|
FBJ murine osteosarcoma viral oncogene homolog B |
| chr22_-_7129631 | 0.50 |
ENSDART00000171359
|
asic1b
|
acid-sensing (proton-gated) ion channel 1b |
| chr14_-_41243180 | 0.49 |
ENSDART00000184548
|
drp2
|
dystrophin related protein 2 |
| chr25_-_21085661 | 0.47 |
ENSDART00000099355
|
prr5a
|
proline rich 5a (renal) |
| chr21_-_28439596 | 0.46 |
ENSDART00000089980
ENSDART00000132844 |
rasgrp2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr23_-_3721444 | 0.46 |
ENSDART00000141682
|
nudt3a
|
nudix (nucleoside diphosphate linked moiety X)-type motif 3a |
| chr15_-_31265375 | 0.46 |
ENSDART00000086592
|
vezf1b
|
vascular endothelial zinc finger 1b |
| chr4_-_9214288 | 0.46 |
ENSDART00000067309
|
avpr1ab
|
arginine vasopressin receptor 1Ab |
| chr12_+_26467847 | 0.45 |
ENSDART00000022495
|
ndel1a
|
nudE neurodevelopment protein 1-like 1a |
| chr15_+_17446796 | 0.43 |
ENSDART00000157189
|
snx19b
|
sorting nexin 19b |
| chr12_-_4475890 | 0.42 |
ENSDART00000092492
|
si:ch211-173d10.1
|
si:ch211-173d10.1 |
| chr3_-_30434016 | 0.42 |
ENSDART00000150958
|
lrrc4ba
|
leucine rich repeat containing 4Ba |
| chr18_-_21806613 | 0.41 |
ENSDART00000145721
|
nrn1la
|
neuritin 1-like a |
| chr7_+_42206847 | 0.40 |
ENSDART00000149250
|
phkb
|
phosphorylase kinase, beta |
| chr4_-_2868112 | 0.40 |
ENSDART00000133843
|
pde3a
|
phosphodiesterase 3A, cGMP-inhibited |
| chr7_-_68419830 | 0.39 |
ENSDART00000191606
|
CU468723.2
|
|
| chr5_-_23675222 | 0.39 |
ENSDART00000135153
|
TBC1D8B
|
si:dkey-110k5.6 |
| chr23_-_17470146 | 0.39 |
ENSDART00000002398
|
trim101
|
tripartite motif containing 101 |
| chr21_-_22812417 | 0.38 |
ENSDART00000079151
ENSDART00000112175 |
trpc6a
|
transient receptor potential cation channel, subfamily C, member 6a |
| chr22_-_7669624 | 0.38 |
ENSDART00000189617
|
AL929276.2
|
|
| chr3_-_8399774 | 0.38 |
ENSDART00000190238
ENSDART00000113140 ENSDART00000132949 |
rbfox3b
|
RNA binding fox-1 homolog 3b |
| chr7_-_71434298 | 0.36 |
ENSDART00000180507
|
lgi2a
|
leucine-rich repeat LGI family, member 2a |
| chr14_-_1313480 | 0.36 |
ENSDART00000097748
|
il21
|
interleukin 21 |
| chr16_+_46492994 | 0.36 |
ENSDART00000134734
|
rpz5
|
rapunzel 5 |
| chr20_+_22130284 | 0.35 |
ENSDART00000025575
|
clocka
|
clock circadian regulator a |
| chr23_-_15216654 | 0.34 |
ENSDART00000131649
|
sulf2b
|
sulfatase 2b |
| chr21_+_20229926 | 0.33 |
ENSDART00000025860
|
si:dkey-247m21.3
|
si:dkey-247m21.3 |
| chr6_+_4255319 | 0.33 |
ENSDART00000170351
|
nbeal1
|
neurobeachin-like 1 |
| chr2_-_31735142 | 0.32 |
ENSDART00000130903
|
ralyl
|
RALY RNA binding protein like |
| chr2_+_42871831 | 0.32 |
ENSDART00000171393
|
efr3a
|
EFR3 homolog A (S. cerevisiae) |
| chr4_+_13810811 | 0.30 |
ENSDART00000067168
|
pdzrn4
|
PDZ domain containing ring finger 4 |
| chr24_-_36680261 | 0.30 |
ENSDART00000059507
|
ccr10
|
chemokine (C-C motif) receptor 10 |
| chr23_+_28770225 | 0.30 |
ENSDART00000132179
ENSDART00000142273 |
masp2
|
mannan-binding lectin serine peptidase 2 |
| chr23_-_11128601 | 0.29 |
ENSDART00000131232
|
cntn3a.2
|
contactin 3a, tandem duplicate 2 |
| chr18_+_36770166 | 0.28 |
ENSDART00000078151
|
fosb
|
FBJ murine osteosarcoma viral oncogene homolog B |
| chr20_-_6812688 | 0.28 |
ENSDART00000170934
|
igfbp1a
|
insulin-like growth factor binding protein 1a |
| chr15_+_11381532 | 0.28 |
ENSDART00000124172
|
si:ch73-321d9.2
|
si:ch73-321d9.2 |
| chr20_+_52546186 | 0.26 |
ENSDART00000110777
ENSDART00000153377 ENSDART00000153013 ENSDART00000042704 |
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr11_-_426525 | 0.26 |
ENSDART00000157054
|
znf831
|
zinc finger protein 831 |
| chr16_-_26255877 | 0.24 |
ENSDART00000146214
|
erfl1
|
Ets2 repressor factor like 1 |
| chr24_+_2962988 | 0.24 |
ENSDART00000164449
|
eci2
|
enoyl-CoA delta isomerase 2 |
| chr1_-_54191626 | 0.24 |
ENSDART00000062941
|
nthl1
|
nth-like DNA glycosylase 1 |
| chr8_+_37168570 | 0.23 |
ENSDART00000098636
|
csf1b
|
colony stimulating factor 1b (macrophage) |
| chr11_+_43114108 | 0.22 |
ENSDART00000013642
ENSDART00000190266 |
foxg1b
|
forkhead box G1b |
| chr8_+_21280360 | 0.21 |
ENSDART00000144488
|
itpr3
|
inositol 1,4,5-trisphosphate receptor, type 3 |
| chr9_+_44430974 | 0.20 |
ENSDART00000056846
|
ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr3_-_32071468 | 0.20 |
ENSDART00000121979
|
BX511080.1
|
|
| chr21_+_3901775 | 0.18 |
ENSDART00000053609
|
dolpp1
|
dolichyldiphosphatase 1 |
| chr16_-_16701718 | 0.18 |
ENSDART00000143550
|
si:dkey-8k3.2
|
si:dkey-8k3.2 |
| chr25_+_32496877 | 0.18 |
ENSDART00000132698
|
ctdspl2a
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2a |
| chr17_+_8175998 | 0.17 |
ENSDART00000131200
|
myct1b
|
myc target 1b |
| chr6_-_49861506 | 0.17 |
ENSDART00000121809
|
gnas
|
GNAS complex locus |
| chr16_-_2650341 | 0.17 |
ENSDART00000128169
ENSDART00000155432 ENSDART00000103722 |
lyplal1
|
lysophospholipase-like 1 |
| chr8_-_14484599 | 0.16 |
ENSDART00000057644
|
lhx4
|
LIM homeobox 4 |
| chr5_+_42136359 | 0.16 |
ENSDART00000083731
|
trpv1
|
transient receptor potential cation channel, subfamily V, member 1 |
| chr15_+_41901970 | 0.15 |
ENSDART00000152724
|
si:ch211-191a16.5
|
si:ch211-191a16.5 |
| chr13_-_24877577 | 0.15 |
ENSDART00000182705
|
kat6b
|
K(lysine) acetyltransferase 6B |
| chr5_-_14211487 | 0.15 |
ENSDART00000135731
|
npffr1l2
|
neuropeptide FF receptor 1 like 2 |
| chr6_-_16456093 | 0.15 |
ENSDART00000083305
ENSDART00000181640 |
slc19a2
|
solute carrier family 19 (thiamine transporter), member 2 |
| chr6_-_7052408 | 0.14 |
ENSDART00000150033
ENSDART00000149232 |
bin1b
|
bridging integrator 1b |
| chr19_-_17526735 | 0.14 |
ENSDART00000189391
|
thrb
|
thyroid hormone receptor beta |
| chr15_-_30984557 | 0.13 |
ENSDART00000080328
|
nf1a
|
neurofibromin 1a |
| chr8_-_621142 | 0.13 |
ENSDART00000187576
ENSDART00000190316 |
si:ch211-87m7.2
|
si:ch211-87m7.2 |
| chr22_-_38360205 | 0.13 |
ENSDART00000162055
|
mark1
|
MAP/microtubule affinity-regulating kinase 1 |
| chr2_+_5563077 | 0.11 |
ENSDART00000111220
|
mb21d2
|
Mab-21 domain containing 2 |
| chr15_-_26552652 | 0.11 |
ENSDART00000152336
|
serpinf2b
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2b |
| chr4_-_13255700 | 0.11 |
ENSDART00000162277
ENSDART00000026593 |
grip1
|
glutamate receptor interacting protein 1 |
| chr4_-_31700186 | 0.11 |
ENSDART00000183986
ENSDART00000180890 |
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr18_+_8937718 | 0.10 |
ENSDART00000138115
|
si:dkey-5i3.1
|
si:dkey-5i3.1 |
| chr23_+_19655301 | 0.09 |
ENSDART00000104441
ENSDART00000135269 |
abhd6b
|
abhydrolase domain containing 6b |
| chr8_-_4010887 | 0.09 |
ENSDART00000163678
|
mtmr3
|
myotubularin related protein 3 |
| chr19_-_32641725 | 0.09 |
ENSDART00000165006
ENSDART00000188185 |
hpca
|
hippocalcin |
| chr7_+_30933867 | 0.08 |
ENSDART00000156325
|
tjp1a
|
tight junction protein 1a |
| chr8_+_35664152 | 0.08 |
ENSDART00000144520
|
si:dkeyp-14d3.1
|
si:dkeyp-14d3.1 |
| chr5_+_7393346 | 0.06 |
ENSDART00000164238
|
si:ch73-72b7.1
|
si:ch73-72b7.1 |
| chr20_-_19422496 | 0.06 |
ENSDART00000143658
|
si:ch211-278j3.3
|
si:ch211-278j3.3 |
| chr5_+_4366431 | 0.06 |
ENSDART00000168560
ENSDART00000149185 |
sat1a.2
|
spermidine/spermine N1-acetyltransferase 1a, duplicate 2 |
| chr19_+_19512515 | 0.04 |
ENSDART00000180065
|
jazf1a
|
JAZF zinc finger 1a |
| chr18_-_17030870 | 0.04 |
ENSDART00000079817
|
has3
|
hyaluronan synthase 3 |
| chr23_-_20002459 | 0.03 |
ENSDART00000163396
|
b4galt3
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 3 |
| chr16_+_22587661 | 0.03 |
ENSDART00000129612
ENSDART00000142241 |
she
|
Src homology 2 domain containing E |
| chr5_-_69212184 | 0.02 |
ENSDART00000053963
|
mat2ab
|
methionine adenosyltransferase II, alpha b |
| chr10_-_7785930 | 0.02 |
ENSDART00000043961
ENSDART00000111058 |
mpx
|
myeloid-specific peroxidase |
| chr22_-_28653074 | 0.02 |
ENSDART00000154717
|
col8a1b
|
collagen, type VIII, alpha 1b |
| chr11_-_22303678 | 0.02 |
ENSDART00000159527
ENSDART00000159681 |
tfeb
|
transcription factor EB |
| chr15_-_39948635 | 0.02 |
ENSDART00000114836
|
msh5
|
mutS homolog 5 |
| chr13_+_28417297 | 0.01 |
ENSDART00000043658
|
slc2a15a
|
solute carrier family 2 (facilitated glucose transporter), member 15a |
| chr6_-_8498908 | 0.01 |
ENSDART00000149222
|
pglyrp2
|
peptidoglycan recognition protein 2 |
| chr10_+_10801719 | 0.00 |
ENSDART00000193648
|
ambp
|
alpha-1-microglobulin/bikunin precursor |
| chr21_+_19525141 | 0.00 |
ENSDART00000058489
|
gzma
|
granzyme A |
Network of associatons between targets according to the STRING database.
First level regulatory network of onecut2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.2 | 0.6 | GO:1903646 | regulation of protein folding(GO:1903332) positive regulation of protein folding(GO:1903334) regulation of chaperone-mediated protein folding(GO:1903644) positive regulation of chaperone-mediated protein folding(GO:1903646) |
| 0.2 | 1.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.2 | 2.0 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.2 | 0.7 | GO:0060300 | microglial cell activation(GO:0001774) regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.2 | 1.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 1.0 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.1 | 0.8 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.1 | 1.3 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 2.1 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 1.8 | GO:0048899 | anterior lateral line development(GO:0048899) |
| 0.1 | 2.4 | GO:0043507 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.1 | 0.5 | GO:1901907 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.5 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.1 | 1.4 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.1 | 0.9 | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902547) |
| 0.1 | 0.4 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 0.5 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 0.4 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 0.2 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.1 | 2.0 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.5 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 2.2 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 0.1 | GO:0046552 | retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.0 | 1.9 | GO:0031103 | axon regeneration(GO:0031103) |
| 0.0 | 0.5 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.7 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.7 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.0 | 0.5 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.8 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 0.2 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.7 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.6 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.1 | GO:0060251 | regulation of glial cell proliferation(GO:0060251) |
| 0.0 | 0.3 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.7 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.3 | GO:0035860 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.3 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.0 | 0.6 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.2 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.8 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.7 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.0 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.1 | GO:0045117 | azole transport(GO:0045117) |
| 0.0 | 0.1 | GO:0006598 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.0 | 0.4 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.2 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.7 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 1.3 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.3 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.3 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.2 | 0.6 | GO:0072380 | TRC complex(GO:0072380) |
| 0.1 | 2.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.6 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.1 | 0.3 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.0 | 0.6 | GO:0033270 | paranode region of axon(GO:0033270) juxtaparanode region of axon(GO:0044224) |
| 0.0 | 1.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.7 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 2.8 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.4 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 1.2 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 2.0 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.5 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 2.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 2.1 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.5 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.2 | GO:0030667 | secretory granule membrane(GO:0030667) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.3 | 2.4 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.2 | 1.4 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.2 | 0.6 | GO:0030882 | lipid antigen binding(GO:0030882) |
| 0.2 | 1.0 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 1.3 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 1.8 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 0.6 | GO:0010858 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.1 | 2.1 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 0.7 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.5 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 0.6 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 0.5 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 1.9 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 0.2 | GO:0000703 | oxidized pyrimidine nucleobase lesion DNA N-glycosylase activity(GO:0000703) |
| 0.1 | 0.6 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 1.2 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 0.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 1.1 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 2.8 | GO:0042562 | hormone binding(GO:0042562) |
| 0.0 | 1.0 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.2 | GO:0050780 | mu-type opioid receptor binding(GO:0031852) dopamine receptor binding(GO:0050780) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.6 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.3 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.1 | GO:0045118 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 1.2 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.8 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.4 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 1.1 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 0.3 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 1.8 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 2.3 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 1.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.8 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.2 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 1.7 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 0.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.1 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 0.5 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 1.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.7 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |