Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
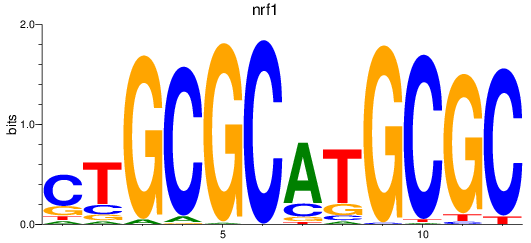
Results for nrf1
Z-value: 3.86
Transcription factors associated with nrf1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nrf1
|
ENSDARG00000000018 | nuclear respiratory factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nrf1 | dr11_v1_chr4_-_15103646_15103696 | 0.73 | 4.1e-04 | Click! |
Activity profile of nrf1 motif
Sorted Z-values of nrf1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_41966854 | 11.38 |
ENSDART00000055709
|
her2
|
hairy-related 2 |
| chr16_-_7379328 | 11.10 |
ENSDART00000060447
|
med18
|
mediator of RNA polymerase II transcription, subunit 18 homolog (yeast) |
| chr24_+_42004640 | 9.73 |
ENSDART00000171380
|
top1mt
|
DNA topoisomerase I mitochondrial |
| chr17_-_20167206 | 9.10 |
ENSDART00000104874
ENSDART00000191995 |
p4ha1b
|
prolyl 4-hydroxylase, alpha polypeptide I b |
| chr20_-_46541834 | 7.91 |
ENSDART00000060685
ENSDART00000181720 |
tmed10
|
transmembrane p24 trafficking protein 10 |
| chr10_-_40826657 | 7.90 |
ENSDART00000076304
|
pcna
|
proliferating cell nuclear antigen |
| chr17_+_24821627 | 7.75 |
ENSDART00000112389
|
wdr43
|
WD repeat domain 43 |
| chr6_-_9676108 | 7.66 |
ENSDART00000169915
|
nop58
|
NOP58 ribonucleoprotein homolog (yeast) |
| chr17_+_37932706 | 7.59 |
ENSDART00000075941
|
plekhh1
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 1 |
| chr16_+_40575742 | 7.27 |
ENSDART00000161503
|
ccne2
|
cyclin E2 |
| chr22_-_16291041 | 7.03 |
ENSDART00000021666
|
rtca
|
RNA 3'-terminal phosphate cyclase |
| chr1_+_25178106 | 6.88 |
ENSDART00000054265
ENSDART00000141648 |
mnd1
|
meiotic nuclear divisions 1 homolog (S. cerevisiae) |
| chr22_+_18319230 | 6.83 |
ENSDART00000184747
ENSDART00000184649 |
gatad2ab
|
GATA zinc finger domain containing 2Ab |
| chr25_+_22320738 | 6.66 |
ENSDART00000073566
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr1_-_25438737 | 6.65 |
ENSDART00000134470
|
fhdc1
|
FH2 domain containing 1 |
| chr17_+_53428092 | 6.45 |
ENSDART00000192509
|
srsf10b
|
serine/arginine-rich splicing factor 10b |
| chr5_-_24882357 | 6.36 |
ENSDART00000127773
ENSDART00000003998 ENSDART00000143851 |
ewsr1b
|
EWS RNA-binding protein 1b |
| chr1_-_21563040 | 6.33 |
ENSDART00000049572
|
ncapd3
|
non-SMC condensin II complex, subunit D3 |
| chr3_+_48473346 | 6.26 |
ENSDART00000166294
|
metrnl
|
meteorin, glial cell differentiation regulator-like |
| chr1_+_260039 | 6.23 |
ENSDART00000092584
ENSDART00000111806 |
cenpe
|
centromere protein E |
| chr24_+_7153628 | 6.19 |
ENSDART00000147715
|
si:dkeyp-101e12.1
|
si:dkeyp-101e12.1 |
| chr21_+_18275961 | 6.12 |
ENSDART00000019750
|
wdr5
|
WD repeat domain 5 |
| chr23_-_18913032 | 6.08 |
ENSDART00000136678
|
si:ch211-209j10.6
|
si:ch211-209j10.6 |
| chr11_+_33818179 | 5.86 |
ENSDART00000109418
|
spoplb
|
speckle-type POZ protein-like b |
| chr20_+_48116476 | 5.79 |
ENSDART00000043938
|
tram2
|
translocation associated membrane protein 2 |
| chr14_+_14806851 | 5.68 |
ENSDART00000169235
|
fhdc2
|
FH2 domain containing 2 |
| chr25_-_12824939 | 5.68 |
ENSDART00000169126
|
uba2
|
ubiquitin-like modifier activating enzyme 2 |
| chr13_-_51903150 | 5.65 |
ENSDART00000090644
|
mrpl2
|
mitochondrial ribosomal protein L2 |
| chr7_+_26534131 | 5.63 |
ENSDART00000173980
|
si:dkey-62k3.5
|
si:dkey-62k3.5 |
| chr17_+_37932433 | 5.62 |
ENSDART00000185349
|
plekhh1
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 1 |
| chr25_-_31629095 | 5.60 |
ENSDART00000170673
ENSDART00000166930 |
lamb1a
|
laminin, beta 1a |
| chr20_-_2725594 | 5.57 |
ENSDART00000152120
|
akirin2
|
akirin 2 |
| chr25_+_22319940 | 5.53 |
ENSDART00000154065
ENSDART00000153492 ENSDART00000024866 ENSDART00000154376 |
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr7_-_20464133 | 5.52 |
ENSDART00000078192
|
cnpy4
|
canopy4 |
| chr17_+_20923691 | 5.50 |
ENSDART00000122407
|
cdk1
|
cyclin-dependent kinase 1 |
| chr5_+_23913585 | 5.36 |
ENSDART00000015401
|
ercc6l
|
excision repair cross-complementation group 6-like |
| chr24_+_35911300 | 5.31 |
ENSDART00000129679
|
abcd4
|
ATP-binding cassette, sub-family D (ALD), member 4 |
| chr1_+_53919110 | 5.31 |
ENSDART00000020680
|
nup133
|
nucleoporin 133 |
| chr18_-_21929426 | 5.27 |
ENSDART00000132034
ENSDART00000079050 |
nutf2
|
nuclear transport factor 2 |
| chr1_+_21563311 | 5.25 |
ENSDART00000147076
ENSDART00000006147 |
primpol
|
primase and polymerase (DNA-directed) |
| chr7_-_41881177 | 5.23 |
ENSDART00000174258
ENSDART00000018972 |
zgc:92818
|
zgc:92818 |
| chr15_-_16384184 | 5.22 |
ENSDART00000154504
|
fam222bb
|
family with sequence similarity 222, member Bb |
| chr16_-_6205790 | 5.21 |
ENSDART00000038495
|
ctnnb1
|
catenin (cadherin-associated protein), beta 1 |
| chr7_-_50367642 | 5.19 |
ENSDART00000134941
|
prc1b
|
protein regulator of cytokinesis 1b |
| chr16_-_21140097 | 5.19 |
ENSDART00000145837
ENSDART00000146500 |
si:dkey-271j15.3
|
si:dkey-271j15.3 |
| chr6_-_9695294 | 5.17 |
ENSDART00000162728
|
nop58
|
NOP58 ribonucleoprotein homolog (yeast) |
| chr3_-_13599482 | 5.17 |
ENSDART00000166639
|
tufm
|
Tu translation elongation factor, mitochondrial |
| chr23_-_44723102 | 5.03 |
ENSDART00000129138
|
mogat3a
|
monoacylglycerol O-acyltransferase 3a |
| chr20_+_25445826 | 4.98 |
ENSDART00000012581
|
pfas
|
phosphoribosylformylglycinamidine synthase |
| chr25_-_35956344 | 4.95 |
ENSDART00000066987
|
mphosph6
|
M-phase phosphoprotein 6 |
| chr14_-_9085349 | 4.92 |
ENSDART00000054710
|
polr1d
|
polymerase (RNA) I polypeptide D |
| chr5_+_60847823 | 4.91 |
ENSDART00000074426
|
lig3
|
ligase III, DNA, ATP-dependent |
| chr12_+_47663419 | 4.91 |
ENSDART00000171932
|
HHEX
|
hematopoietically expressed homeobox |
| chr14_-_26377044 | 4.89 |
ENSDART00000022236
|
emx3
|
empty spiracles homeobox 3 |
| chr16_+_28728347 | 4.89 |
ENSDART00000149240
|
si:dkey-24i24.3
|
si:dkey-24i24.3 |
| chr20_+_34770197 | 4.87 |
ENSDART00000018304
|
mcm3
|
minichromosome maintenance complex component 3 |
| chr7_+_57088920 | 4.86 |
ENSDART00000024076
|
scamp2l
|
secretory carrier membrane protein 2, like |
| chr15_+_26933196 | 4.82 |
ENSDART00000023842
|
ppm1da
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Da |
| chr6_-_8392104 | 4.80 |
ENSDART00000081561
ENSDART00000181178 |
ilf3a
|
interleukin enhancer binding factor 3a |
| chr13_+_25364753 | 4.76 |
ENSDART00000027428
|
chuk
|
conserved helix-loop-helix ubiquitous kinase |
| chr17_-_29224908 | 4.74 |
ENSDART00000156288
|
si:dkey-28g23.6
|
si:dkey-28g23.6 |
| chr6_-_19340889 | 4.63 |
ENSDART00000181407
|
mif4gda
|
MIF4G domain containing a |
| chr20_-_2725930 | 4.62 |
ENSDART00000081643
|
akirin2
|
akirin 2 |
| chr6_+_149405 | 4.60 |
ENSDART00000161154
|
fdx1l
|
ferredoxin 1-like |
| chr11_-_26375575 | 4.59 |
ENSDART00000079255
|
utp3
|
UTP3, small subunit (SSU) processome component, homolog (S. cerevisiae) |
| chr13_+_159504 | 4.58 |
ENSDART00000057248
|
ewsr1b
|
EWS RNA-binding protein 1b |
| chr4_-_9909371 | 4.56 |
ENSDART00000102656
|
si:dkey-22l11.6
|
si:dkey-22l11.6 |
| chr5_+_61459422 | 4.56 |
ENSDART00000050902
|
polr2j
|
polymerase (RNA) II (DNA directed) polypeptide J |
| chr18_-_7448047 | 4.53 |
ENSDART00000193213
ENSDART00000131940 ENSDART00000186944 ENSDART00000052803 |
si:dkey-30c15.10
|
si:dkey-30c15.10 |
| chr12_-_49168398 | 4.53 |
ENSDART00000186608
|
FO704624.1
|
|
| chr9_+_53276356 | 4.52 |
ENSDART00000003310
|
sox21b
|
SRY (sex determining region Y)-box 21b |
| chr22_-_4989803 | 4.52 |
ENSDART00000181359
ENSDART00000125265 ENSDART00000028634 ENSDART00000183294 |
cherp
|
calcium homeostasis endoplasmic reticulum protein |
| chr16_+_52847079 | 4.49 |
ENSDART00000163151
|
cep72
|
centrosomal protein 72 |
| chr21_+_25625026 | 4.49 |
ENSDART00000134678
|
ovol1b
|
ovo-like zinc finger 1b |
| chr5_+_29803380 | 4.48 |
ENSDART00000005263
ENSDART00000137558 ENSDART00000146963 ENSDART00000134900 |
usf1l
|
upstream transcription factor 1, like |
| chr21_-_4849029 | 4.48 |
ENSDART00000168930
ENSDART00000151019 |
notch1a
|
notch 1a |
| chr21_+_43702016 | 4.47 |
ENSDART00000017176
|
dkc1
|
dyskeratosis congenita 1, dyskerin |
| chr5_+_41793001 | 4.42 |
ENSDART00000136439
ENSDART00000190477 ENSDART00000192289 |
bcl7a
|
BCL tumor suppressor 7A |
| chr21_+_17956856 | 4.36 |
ENSDART00000080431
|
dnai1.2
|
dynein, axonemal, intermediate chain 1, paralog 2 |
| chr9_+_32301456 | 4.26 |
ENSDART00000078608
ENSDART00000185153 ENSDART00000144947 |
hspe1
|
heat shock 10 protein 1 |
| chr24_+_4978055 | 4.25 |
ENSDART00000045813
|
zic4
|
zic family member 4 |
| chr1_-_25438934 | 4.25 |
ENSDART00000111686
|
fhdc1
|
FH2 domain containing 1 |
| chr19_-_18127629 | 4.22 |
ENSDART00000187722
|
snx10a
|
sorting nexin 10a |
| chr23_+_29569893 | 4.21 |
ENSDART00000078304
|
lzic
|
leucine zipper and CTNNBIP1 domain containing |
| chr16_-_42461263 | 4.19 |
ENSDART00000109259
|
smarcc1a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1a |
| chr1_-_42289704 | 4.19 |
ENSDART00000150124
|
si:ch211-71k14.1
|
si:ch211-71k14.1 |
| chr22_-_7050 | 4.18 |
ENSDART00000127829
|
atad3
|
ATPase family, AAA domain containing 3 |
| chr21_+_44684577 | 4.17 |
ENSDART00000099528
|
spry4
|
sprouty homolog 4 (Drosophila) |
| chr25_-_12824656 | 4.16 |
ENSDART00000171801
|
uba2
|
ubiquitin-like modifier activating enzyme 2 |
| chr24_-_7957581 | 4.15 |
ENSDART00000145815
|
txndc5
|
thioredoxin domain containing 5 |
| chr10_+_31244619 | 4.15 |
ENSDART00000145562
ENSDART00000184412 |
robo4
|
roundabout, axon guidance receptor, homolog 4 (Drosophila) |
| chr23_+_10805188 | 4.14 |
ENSDART00000035693
|
ppp4r2a
|
protein phosphatase 4, regulatory subunit 2a |
| chr5_+_1493767 | 4.14 |
ENSDART00000022132
|
haus4
|
HAUS augmin-like complex, subunit 4 |
| chr15_-_14884332 | 4.12 |
ENSDART00000165237
|
si:ch211-24o8.4
|
si:ch211-24o8.4 |
| chr12_-_34435604 | 4.12 |
ENSDART00000115088
|
birc5a
|
baculoviral IAP repeat containing 5a |
| chr5_+_36666715 | 4.12 |
ENSDART00000097686
|
zgc:153990
|
zgc:153990 |
| chr22_+_18319666 | 4.11 |
ENSDART00000033103
|
gatad2ab
|
GATA zinc finger domain containing 2Ab |
| chr11_+_31236001 | 4.10 |
ENSDART00000129393
|
trmt1
|
tRNA methyltransferase 1 |
| chr4_-_25858620 | 4.08 |
ENSDART00000128968
|
nr2c1
|
nuclear receptor subfamily 2, group C, member 1 |
| chr19_-_18127808 | 4.08 |
ENSDART00000108627
|
snx10a
|
sorting nexin 10a |
| chr24_+_4977862 | 4.06 |
ENSDART00000114537
|
zic4
|
zic family member 4 |
| chr18_+_46382484 | 4.04 |
ENSDART00000024202
ENSDART00000142790 |
daw1
|
dynein assembly factor with WDR repeat domains 1 |
| chr16_-_32221312 | 4.02 |
ENSDART00000112348
|
calhm5.1
|
calcium homeostasis modulator family member 5, tandem duplicate 1 |
| chr4_-_11024767 | 3.95 |
ENSDART00000067261
ENSDART00000167631 |
tmtc2a
|
transmembrane and tetratricopeptide repeat containing 2a |
| chr25_-_35296165 | 3.95 |
ENSDART00000018107
|
fancf
|
Fanconi anemia, complementation group F |
| chr21_+_15601100 | 3.95 |
ENSDART00000180558
ENSDART00000145454 |
smarcb1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1b |
| chr19_+_24891747 | 3.94 |
ENSDART00000132209
ENSDART00000193610 |
eya3
|
EYA transcriptional coactivator and phosphatase 3 |
| chr14_-_21123325 | 3.94 |
ENSDART00000180889
ENSDART00000188539 |
si:dkey-74k8.4
|
si:dkey-74k8.4 |
| chr5_+_50869091 | 3.90 |
ENSDART00000083294
|
nol6
|
nucleolar protein 6 (RNA-associated) |
| chr6_+_56141852 | 3.89 |
ENSDART00000149665
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr6_-_19341184 | 3.89 |
ENSDART00000168236
ENSDART00000167674 |
mif4gda
|
MIF4G domain containing a |
| chr8_-_14239023 | 3.88 |
ENSDART00000090371
|
sin3b
|
SIN3 transcription regulator family member B |
| chr17_+_28706946 | 3.87 |
ENSDART00000126967
|
strn3
|
striatin, calmodulin binding protein 3 |
| chr2_+_25839650 | 3.85 |
ENSDART00000134077
ENSDART00000140804 |
eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr20_+_50015470 | 3.82 |
ENSDART00000189319
|
riox1
|
ribosomal oxygenase 1 |
| chr5_+_19933356 | 3.80 |
ENSDART00000088819
|
ankrd13a
|
ankyrin repeat domain 13A |
| chr2_+_29996650 | 3.79 |
ENSDART00000138050
ENSDART00000141026 |
rbm33b
|
RNA binding motif protein 33b |
| chr24_-_38657683 | 3.77 |
ENSDART00000154843
|
si:ch1073-164k15.3
|
si:ch1073-164k15.3 |
| chr13_-_35844961 | 3.77 |
ENSDART00000171371
|
map3k4
|
mitogen-activated protein kinase kinase kinase 4 |
| chr25_-_17852944 | 3.75 |
ENSDART00000155169
|
si:ch211-176l24.4
|
si:ch211-176l24.4 |
| chr21_+_18274825 | 3.73 |
ENSDART00000144322
ENSDART00000147768 |
wdr5
|
WD repeat domain 5 |
| chr19_-_48039400 | 3.69 |
ENSDART00000166748
ENSDART00000165921 |
csf3b
|
colony stimulating factor 3 (granulocyte) b |
| chr16_-_4769877 | 3.66 |
ENSDART00000149421
ENSDART00000054078 |
rpa2
|
replication protein A2 |
| chr22_-_24285432 | 3.64 |
ENSDART00000164083
|
si:ch211-117l17.4
|
si:ch211-117l17.4 |
| chr16_-_4770233 | 3.63 |
ENSDART00000193228
|
rpa2
|
replication protein A2 |
| chr13_+_25364324 | 3.62 |
ENSDART00000187471
|
chuk
|
conserved helix-loop-helix ubiquitous kinase |
| chr14_+_24277556 | 3.62 |
ENSDART00000122660
|
hnrnpa0a
|
heterogeneous nuclear ribonucleoprotein A0a |
| chr10_+_19026988 | 3.61 |
ENSDART00000137456
|
igsf9a
|
immunoglobulin superfamily, member 9a |
| chr22_-_15704704 | 3.59 |
ENSDART00000017838
ENSDART00000130238 |
safb
|
scaffold attachment factor B |
| chr22_+_25931782 | 3.59 |
ENSDART00000157842
|
dnaja3b
|
DnaJ (Hsp40) homolog, subfamily A, member 3B |
| chr15_+_24549054 | 3.57 |
ENSDART00000155900
|
phf12b
|
PHD finger protein 12b |
| chr7_+_57089354 | 3.56 |
ENSDART00000140702
|
scamp2l
|
secretory carrier membrane protein 2, like |
| chr22_+_336256 | 3.56 |
ENSDART00000019155
|
btg2
|
B-cell translocation gene 2 |
| chr13_+_5004924 | 3.56 |
ENSDART00000056295
ENSDART00000130379 |
psap
|
prosaposin |
| chr25_+_2776511 | 3.54 |
ENSDART00000115280
|
neo1b
|
neogenin 1b |
| chr7_-_50367326 | 3.53 |
ENSDART00000141926
|
prc1b
|
protein regulator of cytokinesis 1b |
| chr8_+_13064750 | 3.52 |
ENSDART00000039878
|
sap30bp
|
SAP30 binding protein |
| chr14_-_1355544 | 3.51 |
ENSDART00000060417
|
cetn4
|
centrin 4 |
| chr18_-_50799510 | 3.50 |
ENSDART00000174373
|
taldo1
|
transaldolase 1 |
| chr21_-_18275226 | 3.50 |
ENSDART00000126672
ENSDART00000135239 |
brd3a
|
bromodomain containing 3a |
| chr17_-_53348486 | 3.49 |
ENSDART00000161183
|
CABZ01066926.1
|
|
| chr13_+_15701849 | 3.48 |
ENSDART00000003517
|
trmt61a
|
tRNA methyltransferase 61A |
| chr19_+_20787179 | 3.47 |
ENSDART00000193204
|
adnp2b
|
ADNP homeobox 2b |
| chr16_+_4770266 | 3.46 |
ENSDART00000038036
|
ube3d
|
ubiquitin protein ligase E3D |
| chr23_+_31815423 | 3.46 |
ENSDART00000075730
ENSDART00000075726 |
myb
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr22_+_37631034 | 3.46 |
ENSDART00000159016
ENSDART00000193346 |
psmd1
|
proteasome 26S subunit, non-ATPase 1 |
| chr19_-_35450661 | 3.45 |
ENSDART00000113574
ENSDART00000136895 |
anln
|
anillin, actin binding protein |
| chr5_-_33281046 | 3.43 |
ENSDART00000051344
ENSDART00000138116 |
surf6
|
surfeit 6 |
| chr1_-_44484 | 3.43 |
ENSDART00000171547
ENSDART00000164075 ENSDART00000168091 |
tmem39a
|
transmembrane protein 39A |
| chr25_-_35101673 | 3.41 |
ENSDART00000140864
|
zgc:162611
|
zgc:162611 |
| chr13_-_36621926 | 3.41 |
ENSDART00000057155
|
cdkn3
|
cyclin-dependent kinase inhibitor 3 |
| chr15_+_16886196 | 3.40 |
ENSDART00000139296
ENSDART00000049196 |
gdpd1
|
glycerophosphodiester phosphodiesterase domain containing 1 |
| chr6_+_48862 | 3.38 |
ENSDART00000082954
|
mbd5
|
methyl-CpG binding domain protein 5 |
| chr2_+_30182431 | 3.36 |
ENSDART00000004903
|
rdh10b
|
retinol dehydrogenase 10b |
| chr20_-_54245256 | 3.36 |
ENSDART00000170482
|
prpf39
|
PRP39 pre-mRNA processing factor 39 homolog (yeast) |
| chr11_-_12233 | 3.35 |
ENSDART00000173352
ENSDART00000173009 ENSDART00000102293 |
myg1
|
melanocyte proliferating gene 1 |
| chr11_-_309420 | 3.35 |
ENSDART00000173185
|
poc1a
|
POC1 centriolar protein A |
| chr2_+_25839193 | 3.35 |
ENSDART00000078634
|
eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr13_+_15702142 | 3.32 |
ENSDART00000135960
|
trmt61a
|
tRNA methyltransferase 61A |
| chr3_+_43086548 | 3.29 |
ENSDART00000163579
|
si:dkey-43p13.5
|
si:dkey-43p13.5 |
| chr7_+_39738505 | 3.28 |
ENSDART00000004365
|
tada2b
|
transcriptional adaptor 2B |
| chr25_+_7532627 | 3.28 |
ENSDART00000187660
|
ptdss2
|
phosphatidylserine synthase 2 |
| chr19_-_30800004 | 3.27 |
ENSDART00000128560
ENSDART00000045504 ENSDART00000125893 |
trit1
|
tRNA isopentenyltransferase 1 |
| chr5_-_32445835 | 3.25 |
ENSDART00000170919
|
ncs1a
|
neuronal calcium sensor 1a |
| chr12_+_10443785 | 3.25 |
ENSDART00000029133
|
snu13b
|
SNU13 homolog, small nuclear ribonucleoprotein b (U4/U6.U5) |
| chr23_-_493384 | 3.25 |
ENSDART00000128433
|
hm13
|
histocompatibility (minor) 13 |
| chr10_-_33265566 | 3.23 |
ENSDART00000063642
|
tbl2
|
transducin (beta)-like 2 |
| chr22_-_25078650 | 3.22 |
ENSDART00000131811
ENSDART00000141546 |
kat14
|
lysine acetyltransferase 14 |
| chr10_+_44984946 | 3.21 |
ENSDART00000182520
|
h2afvb
|
H2A histone family, member Vb |
| chr16_-_7793457 | 3.20 |
ENSDART00000113483
|
trim71
|
tripartite motif containing 71, E3 ubiquitin protein ligase |
| chr25_-_35101396 | 3.20 |
ENSDART00000138865
|
zgc:162611
|
zgc:162611 |
| chr15_+_9861973 | 3.19 |
ENSDART00000170945
|
si:dkey-13m3.2
|
si:dkey-13m3.2 |
| chr11_-_36450770 | 3.19 |
ENSDART00000128889
|
zbtb40
|
zinc finger and BTB domain containing 40 |
| chr16_-_30563129 | 3.17 |
ENSDART00000191716
|
lmna
|
lamin A |
| chr5_-_41142467 | 3.16 |
ENSDART00000129415
|
zfr
|
zinc finger RNA binding protein |
| chr21_-_34032650 | 3.16 |
ENSDART00000138575
ENSDART00000047515 |
rnf145b
|
ring finger protein 145b |
| chr1_-_39983730 | 3.15 |
ENSDART00000160066
|
ing2
|
inhibitor of growth family, member 2 |
| chr4_+_966061 | 3.14 |
ENSDART00000122535
|
rpap3
|
RNA polymerase II associated protein 3 |
| chr15_-_28587147 | 3.14 |
ENSDART00000156049
|
ssh2a
|
slingshot protein phosphatase 2a |
| chr19_-_25693728 | 3.12 |
ENSDART00000136187
|
bloc1s4
|
biogenesis of lysosomal organelles complex-1, subunit 4, cappuccino |
| chr5_-_69716501 | 3.10 |
ENSDART00000158956
|
mob1a
|
MOB kinase activator 1A |
| chr24_-_24168475 | 3.10 |
ENSDART00000139369
ENSDART00000080671 |
eif1axb
|
eukaryotic translation initiation factor 1A, X-linked, b |
| chr3_+_15736811 | 3.09 |
ENSDART00000080075
|
phb
|
prohibitin |
| chr23_+_26079467 | 3.09 |
ENSDART00000129617
|
atp6ap1b
|
ATPase H+ transporting accessory protein 1b |
| chr25_+_30460378 | 3.07 |
ENSDART00000016310
|
banp
|
BTG3 associated nuclear protein |
| chr19_-_42588510 | 3.05 |
ENSDART00000102583
|
sytl1
|
synaptotagmin-like 1 |
| chr18_-_7097403 | 3.05 |
ENSDART00000003748
|
cfap161
|
cilia and flagella associated protein 161 |
| chr20_-_51547464 | 3.03 |
ENSDART00000099486
|
disp1
|
dispatched homolog 1 (Drosophila) |
| chr5_-_19932621 | 3.03 |
ENSDART00000088881
|
git2a
|
G protein-coupled receptor kinase interacting ArfGAP 2a |
| chr3_+_26342768 | 3.00 |
ENSDART00000163832
|
si:ch211-156b7.4
|
si:ch211-156b7.4 |
| chr11_+_12744575 | 2.99 |
ENSDART00000131059
ENSDART00000081335 ENSDART00000142481 |
rbb4l
|
retinoblastoma binding protein 4, like |
| chr21_+_249970 | 2.99 |
ENSDART00000169026
|
jak2a
|
Janus kinase 2a |
| chr14_-_26411918 | 2.98 |
ENSDART00000020582
|
tmed9
|
transmembrane p24 trafficking protein 9 |
| chr3_+_38540411 | 2.96 |
ENSDART00000154943
|
si:dkey-7f16.3
|
si:dkey-7f16.3 |
| chr22_+_15343953 | 2.95 |
ENSDART00000045682
|
rrp36
|
ribosomal RNA processing 36 |
| chr12_+_27022517 | 2.95 |
ENSDART00000152975
|
msl1b
|
male-specific lethal 1 homolog b (Drosophila) |
| chr21_+_37090585 | 2.94 |
ENSDART00000182971
|
znf346
|
zinc finger protein 346 |
| chr22_+_37631234 | 2.91 |
ENSDART00000007346
|
psmd1
|
proteasome 26S subunit, non-ATPase 1 |
| chr3_-_37082618 | 2.91 |
ENSDART00000026701
ENSDART00000110716 |
tubg1
|
tubulin, gamma 1 |
| chr8_-_51954562 | 2.90 |
ENSDART00000132527
ENSDART00000057315 |
cep78
|
centrosomal protein 78 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nrf1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 10.2 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 2.4 | 12.2 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 2.0 | 7.9 | GO:0006272 | leading strand elongation(GO:0006272) |
| 1.9 | 11.4 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 1.8 | 5.5 | GO:1903392 | epicardial cell to mesenchymal cell transition(GO:0003347) negative regulation of adherens junction organization(GO:1903392) |
| 1.7 | 5.2 | GO:2000055 | positive regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000055) |
| 1.7 | 5.2 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 1.7 | 10.1 | GO:0055016 | hypochord development(GO:0055016) |
| 1.7 | 8.4 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 1.7 | 8.3 | GO:0070986 | left/right axis specification(GO:0070986) |
| 1.6 | 6.3 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 1.5 | 4.5 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 1.5 | 10.3 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 1.4 | 4.1 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 1.4 | 4.1 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 1.3 | 9.2 | GO:0045901 | positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 1.3 | 3.9 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 1.2 | 5.0 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 1.2 | 7.4 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 1.2 | 8.5 | GO:0032185 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 1.2 | 8.3 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 1.1 | 5.7 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 1.1 | 5.5 | GO:0045448 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 1.1 | 3.2 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 1.1 | 6.3 | GO:0090342 | regulation of cell aging(GO:0090342) |
| 1.1 | 4.2 | GO:0070920 | regulation of production of small RNA involved in gene silencing by RNA(GO:0070920) regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903798) |
| 1.0 | 2.0 | GO:1900364 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 1.0 | 3.0 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 1.0 | 2.9 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 1.0 | 3.8 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.9 | 10.0 | GO:0000022 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.9 | 3.5 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.9 | 2.6 | GO:1905048 | regulation of metallopeptidase activity(GO:1905048) |
| 0.9 | 2.6 | GO:1904983 | transmembrane glycine transport from cytosol to mitochondrion(GO:1904983) |
| 0.8 | 3.4 | GO:0070291 | N-acylethanolamine metabolic process(GO:0070291) |
| 0.8 | 2.5 | GO:0009193 | ribonucleoside diphosphate catabolic process(GO:0009191) pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.8 | 4.9 | GO:1902975 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.8 | 5.6 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.8 | 5.3 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.7 | 2.2 | GO:0072526 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 0.7 | 3.5 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.7 | 4.8 | GO:0046294 | formaldehyde metabolic process(GO:0046292) formaldehyde catabolic process(GO:0046294) |
| 0.7 | 6.1 | GO:2000758 | positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.7 | 4.6 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.6 | 2.5 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.6 | 10.5 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.6 | 3.1 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.6 | 2.5 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.6 | 11.7 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.6 | 6.2 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.6 | 5.8 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.6 | 3.5 | GO:0045658 | regulation of granulocyte differentiation(GO:0030852) regulation of neutrophil differentiation(GO:0045658) |
| 0.6 | 3.4 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.5 | 3.7 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.5 | 2.1 | GO:0006404 | RNA import into nucleus(GO:0006404) snRNA import into nucleus(GO:0061015) |
| 0.5 | 4.6 | GO:1901099 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.5 | 5.6 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.5 | 5.0 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.5 | 1.5 | GO:0051958 | folic acid transport(GO:0015884) drug transport(GO:0015893) methotrexate transport(GO:0051958) reduced folate transmembrane transport(GO:0098838) |
| 0.5 | 2.5 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.5 | 5.0 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.5 | 6.4 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.5 | 2.4 | GO:0045056 | transcytosis(GO:0045056) |
| 0.5 | 1.4 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.5 | 6.1 | GO:0014034 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.5 | 2.3 | GO:0060760 | positive regulation of cytokine-mediated signaling pathway(GO:0001961) positive regulation of response to cytokine stimulus(GO:0060760) |
| 0.5 | 3.2 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.5 | 2.3 | GO:0060368 | regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) |
| 0.4 | 2.7 | GO:0003342 | proepicardium development(GO:0003342) septum transversum development(GO:0003343) determination of intestine left/right asymmetry(GO:0071908) |
| 0.4 | 1.8 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.4 | 13.2 | GO:0001569 | patterning of blood vessels(GO:0001569) |
| 0.4 | 2.5 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.4 | 3.9 | GO:0043931 | ossification involved in bone maturation(GO:0043931) bone maturation(GO:0070977) |
| 0.4 | 1.9 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.4 | 2.3 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.4 | 2.3 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.4 | 6.1 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.4 | 3.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.4 | 8.8 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.3 | 1.6 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.3 | 1.6 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.3 | 8.5 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.3 | 9.1 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.3 | 0.9 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.3 | 2.1 | GO:0070285 | pigment cell development(GO:0070285) |
| 0.3 | 1.5 | GO:0060544 | regulation of necrotic cell death(GO:0010939) regulation of necroptotic process(GO:0060544) |
| 0.3 | 5.4 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.3 | 9.6 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.3 | 2.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.3 | 2.7 | GO:0032986 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.3 | 8.3 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.3 | 2.9 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.3 | 2.6 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.3 | 4.0 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.3 | 2.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.3 | 0.8 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.3 | 0.8 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 0.3 | 3.4 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.3 | 2.5 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.3 | 1.5 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.2 | 2.0 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.2 | 1.2 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.2 | 1.7 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.2 | 3.0 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.2 | 1.8 | GO:0046149 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.2 | 1.1 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.2 | 1.8 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.2 | 3.7 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.2 | 3.5 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.2 | 1.1 | GO:0072103 | renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.2 | 0.8 | GO:0090085 | regulation of protein deubiquitination(GO:0090085) |
| 0.2 | 4.6 | GO:0009408 | response to heat(GO:0009408) |
| 0.2 | 1.3 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.2 | 0.4 | GO:0045649 | regulation of macrophage differentiation(GO:0045649) |
| 0.2 | 2.5 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.2 | 6.3 | GO:0006289 | nucleotide-excision repair(GO:0006289) |
| 0.2 | 1.6 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.2 | 1.0 | GO:1903426 | regulation of reactive oxygen species biosynthetic process(GO:1903426) |
| 0.2 | 4.1 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.2 | 4.9 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.2 | 1.5 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) |
| 0.2 | 2.0 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.2 | 1.3 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.2 | 4.3 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.2 | 8.4 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.2 | 5.7 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.2 | 9.0 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.2 | 12.4 | GO:0034504 | protein localization to nucleus(GO:0034504) |
| 0.2 | 6.0 | GO:0060872 | semicircular canal development(GO:0060872) |
| 0.2 | 3.8 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.2 | 2.2 | GO:0070189 | kynurenine metabolic process(GO:0070189) |
| 0.2 | 1.6 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.2 | 3.9 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.2 | 2.0 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.2 | 3.7 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.2 | 6.1 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) |
| 0.1 | 5.0 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.1 | 10.2 | GO:0045089 | positive regulation of innate immune response(GO:0045089) |
| 0.1 | 35.1 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.1 | 1.3 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.1 | 2.9 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 3.0 | GO:0046427 | positive regulation of JAK-STAT cascade(GO:0046427) positive regulation of STAT cascade(GO:1904894) |
| 0.1 | 2.7 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.1 | 1.9 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.1 | 6.3 | GO:0055072 | iron ion homeostasis(GO:0055072) |
| 0.1 | 2.0 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 0.7 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 11.0 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.1 | 2.6 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.1 | 2.2 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.1 | 1.0 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 2.7 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.1 | 0.7 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 0.2 | GO:0002532 | production of molecular mediator involved in inflammatory response(GO:0002532) |
| 0.1 | 0.6 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.1 | 3.4 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.1 | 4.2 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.1 | 5.8 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 4.1 | GO:0043523 | regulation of neuron apoptotic process(GO:0043523) |
| 0.1 | 3.7 | GO:0021954 | central nervous system neuron development(GO:0021954) |
| 0.1 | 1.4 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.1 | 2.2 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 21.1 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.1 | 1.0 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 4.2 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.1 | 1.5 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.1 | 2.0 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.1 | 3.3 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.1 | 2.0 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.1 | 1.3 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 3.0 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.1 | 1.1 | GO:0030878 | thyroid gland development(GO:0030878) |
| 0.1 | 0.4 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.1 | 1.8 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 1.5 | GO:0006378 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.1 | 0.9 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 3.6 | GO:0050684 | regulation of mRNA processing(GO:0050684) |
| 0.1 | 2.9 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.1 | 2.5 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 | 0.6 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.1 | 3.6 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.1 | 2.9 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.1 | 4.8 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.1 | 0.3 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 1.9 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.1 | 1.3 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 1.1 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.4 | GO:0030719 | P granule organization(GO:0030719) |
| 0.0 | 0.7 | GO:0055117 | regulation of cardiac muscle contraction(GO:0055117) |
| 0.0 | 0.3 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 0.5 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 | 1.1 | GO:0071230 | positive regulation of TOR signaling(GO:0032008) cellular response to amino acid stimulus(GO:0071230) |
| 0.0 | 1.1 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 1.4 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 2.8 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 1.3 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 4.0 | GO:0009913 | epidermal cell differentiation(GO:0009913) |
| 0.0 | 3.0 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.0 | 3.2 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.6 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 1.5 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.4 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.0 | 1.6 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.5 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 3.6 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.0 | 0.4 | GO:0015800 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.4 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.2 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.6 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.5 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 3.7 | GO:0042176 | regulation of protein catabolic process(GO:0042176) |
| 0.0 | 0.4 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.4 | GO:0070828 | heterochromatin organization(GO:0070828) |
| 0.0 | 0.7 | GO:0048634 | regulation of muscle organ development(GO:0048634) |
| 0.0 | 0.9 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 2.5 | GO:0001756 | somitogenesis(GO:0001756) |
| 0.0 | 1.6 | GO:0030100 | regulation of endocytosis(GO:0030100) |
| 0.0 | 1.4 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.7 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 3.3 | GO:0051604 | protein maturation(GO:0051604) |
| 0.0 | 9.9 | GO:0006396 | RNA processing(GO:0006396) |
| 0.0 | 0.9 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 1.0 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 1.2 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 2.8 | GO:0007059 | chromosome segregation(GO:0007059) |
| 0.0 | 8.2 | GO:0006325 | chromatin organization(GO:0006325) |
| 0.0 | 4.1 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0040001 | establishment of mitotic spindle localization(GO:0040001) |
| 0.0 | 1.7 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 1.0 | GO:0007265 | Ras protein signal transduction(GO:0007265) |
| 0.0 | 0.4 | GO:0034599 | cellular response to oxidative stress(GO:0034599) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 1.8 | GO:0050770 | regulation of axonogenesis(GO:0050770) |
| 0.0 | 0.2 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 0.6 | GO:0044344 | fibroblast growth factor receptor signaling pathway(GO:0008543) cellular response to fibroblast growth factor stimulus(GO:0044344) response to fibroblast growth factor(GO:0071774) |
| 0.0 | 0.5 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 16.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 2.5 | 10.0 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 2.5 | 9.8 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 2.1 | 6.4 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 1.8 | 12.8 | GO:0070847 | core mediator complex(GO:0070847) |
| 1.4 | 8.4 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 1.4 | 4.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 1.4 | 6.8 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 1.3 | 3.9 | GO:0034456 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 1.2 | 8.5 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 1.2 | 8.5 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 1.1 | 4.5 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.9 | 5.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.8 | 3.2 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.7 | 5.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.7 | 2.2 | GO:0008352 | katanin complex(GO:0008352) |
| 0.7 | 10.8 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.7 | 8.7 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.7 | 4.7 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.7 | 5.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.6 | 2.6 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.6 | 10.6 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.6 | 5.3 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.6 | 10.9 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.6 | 6.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.5 | 1.6 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.5 | 1.6 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.5 | 10.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.5 | 3.1 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.5 | 1.5 | GO:0097361 | CIA complex(GO:0097361) |
| 0.5 | 5.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.5 | 4.6 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.5 | 4.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.4 | 2.6 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.4 | 4.4 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.4 | 2.5 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.4 | 3.9 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.4 | 3.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.3 | 3.4 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.3 | 7.5 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.3 | 2.3 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.3 | 7.5 | GO:0071004 | U2-type prespliceosome(GO:0071004) prespliceosome(GO:0071010) |
| 0.3 | 2.5 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.3 | 2.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.3 | 12.4 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.3 | 1.5 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.3 | 1.6 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.3 | 4.8 | GO:0070461 | SAGA-type complex(GO:0070461) |
| 0.3 | 2.9 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.3 | 15.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.3 | 1.5 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.3 | 5.6 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.3 | 3.8 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.2 | 3.7 | GO:0044545 | NSL complex(GO:0044545) |
| 0.2 | 3.5 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.2 | 2.0 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.2 | 1.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.2 | 4.1 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.2 | 1.9 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.2 | 3.0 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 14.5 | GO:0000779 | condensed chromosome, centromeric region(GO:0000779) |
| 0.2 | 3.0 | GO:0045180 | basal cortex(GO:0045180) |
| 0.2 | 13.4 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.2 | 2.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.2 | 5.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 3.1 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 0.6 | GO:0010369 | chromocenter(GO:0010369) |
| 0.2 | 2.8 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.2 | 0.9 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.2 | 8.1 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.2 | 1.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.2 | 3.7 | GO:0032039 | integrator complex(GO:0032039) |
| 0.2 | 11.8 | GO:0030684 | preribosome(GO:0030684) |
| 0.2 | 7.0 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.2 | 2.6 | GO:0036038 | MKS complex(GO:0036038) |
| 0.2 | 1.3 | GO:0034464 | BBSome(GO:0034464) |
| 0.2 | 2.7 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.2 | 1.3 | GO:0032797 | SMN complex(GO:0032797) |
| 0.2 | 1.3 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 0.2 | 1.7 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 2.1 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.1 | 3.4 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 2.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 2.0 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 5.3 | GO:0005657 | replication fork(GO:0005657) |
| 0.1 | 3.0 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 0.5 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 10.2 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 1.0 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 2.8 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 1.0 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 2.3 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.1 | 1.2 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.1 | 0.4 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 13.8 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.5 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 10.0 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 8.2 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.8 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 2.9 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.8 | GO:0044447 | axoneme part(GO:0044447) |
| 0.0 | 2.6 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 2.3 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 5.9 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 4.0 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 9.0 | GO:0019866 | mitochondrial inner membrane(GO:0005743) organelle inner membrane(GO:0019866) |
| 0.0 | 0.2 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.9 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 2.4 | GO:0005774 | vacuolar membrane(GO:0005774) |
| 0.0 | 0.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 1.5 | GO:0030117 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 21.9 | GO:0005829 | cytosol(GO:0005829) |
| 0.0 | 0.6 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 2.1 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 1.5 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.8 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 12.6 | GO:0044428 | nuclear part(GO:0044428) |
| 0.0 | 46.2 | GO:0005634 | nucleus(GO:0005634) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 12.2 | GO:0016713 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 2.9 | 11.4 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 2.5 | 9.8 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 2.4 | 11.9 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 2.1 | 8.4 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 1.4 | 6.8 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 1.0 | 4.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 1.0 | 1.9 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.9 | 8.5 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.9 | 10.9 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.9 | 5.4 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.8 | 2.3 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.7 | 2.2 | GO:0047804 | cysteine-S-conjugate beta-lyase activity(GO:0047804) |
| 0.7 | 2.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.6 | 1.9 | GO:0000035 | acyl binding(GO:0000035) |
| 0.6 | 3.9 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.6 | 2.6 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.6 | 12.8 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.6 | 2.5 | GO:1990931 | RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.6 | 4.3 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.6 | 16.5 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.6 | 8.8 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.6 | 3.5 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.6 | 5.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.6 | 5.6 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.6 | 5.0 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.6 | 5.5 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.5 | 5.5 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.5 | 2.2 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.5 | 6.0 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.5 | 1.6 | GO:0004394 | heparan sulfate 2-O-sulfotransferase activity(GO:0004394) |
| 0.5 | 2.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.5 | 2.6 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.5 | 5.0 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.5 | 2.0 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.5 | 1.5 | GO:0008518 | reduced folate carrier activity(GO:0008518) methotrexate transporter activity(GO:0015350) |
| 0.5 | 2.5 | GO:0019863 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.5 | 2.7 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.4 | 2.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.4 | 1.7 | GO:0018812 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.4 | 1.2 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.4 | 3.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.4 | 1.6 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.4 | 1.5 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.4 | 1.1 | GO:1902945 | metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902945) |
| 0.4 | 4.6 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.4 | 5.0 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.3 | 1.7 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.3 | 3.4 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.3 | 6.3 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.3 | 4.1 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.3 | 3.5 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.3 | 0.9 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.3 | 3.7 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.3 | 10.8 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.3 | 17.5 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.3 | 2.6 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.3 | 2.8 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.3 | 1.6 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.3 | 3.6 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.3 | 2.0 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.2 | 4.8 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.2 | 4.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.2 | 6.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.2 | 3.8 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.2 | 2.9 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.2 | 9.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 1.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.2 | 2.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 4.9 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.2 | 3.0 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.2 | 3.6 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.2 | 1.5 | GO:0051219 | protein phosphorylated amino acid binding(GO:0045309) phosphoprotein binding(GO:0051219) |
| 0.2 | 2.6 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.2 | 1.6 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.2 | 4.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.2 | 2.7 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.2 | 0.5 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.2 | 3.4 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.2 | 2.6 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.2 | 6.7 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.2 | 4.7 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.2 | 2.5 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.2 | 1.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 2.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 2.5 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 9.0 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 1.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 5.1 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 0.7 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 7.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 16.6 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 51.3 | GO:0003712 | transcription factor activity, transcription factor binding(GO:0000989) transcription cofactor activity(GO:0003712) |
| 0.1 | 4.2 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 2.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.8 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 0.3 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 2.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 4.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 0.4 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.1 | 1.9 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 2.1 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 2.1 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 7.3 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 3.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 6.6 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.1 | 4.8 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.1 | 0.8 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 2.1 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 1.9 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 1.7 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 0.7 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.1 | 1.0 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 14.7 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 0.6 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 6.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 1.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.6 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.1 | 1.8 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 5.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 0.3 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 7.1 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.1 | 0.2 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 1.4 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 2.6 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 2.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 1.4 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 0.4 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 0.8 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.5 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 10.5 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 1.8 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 42.7 | GO:0003723 | RNA binding(GO:0003723) |
| 0.0 | 1.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 1.1 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.0 | 2.3 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.6 | GO:0019209 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.0 | 1.4 | GO:0098632 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.7 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.0 | 1.2 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.8 | GO:0016655 | oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor(GO:0016655) |
| 0.0 | 0.4 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.9 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.9 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.4 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 1.0 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 1.3 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.0 | 0.9 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 1.7 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.6 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 3.8 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 1.4 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 51.4 | GO:0003676 | nucleic acid binding(GO:0003676) |
| 0.0 | 0.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 1.7 | GO:0004713 | protein tyrosine kinase activity(GO:0004713) |
| 0.0 | 2.6 | GO:0005125 | cytokine activity(GO:0005125) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 10.0 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.5 | 5.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.4 | 12.0 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.4 | 11.8 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.4 | 16.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.3 | 5.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.3 | 8.5 | PID ATR PATHWAY | ATR signaling pathway |
| 0.2 | 3.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.2 | 2.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.2 | 3.8 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.2 | 6.8 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.2 | 8.3 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.2 | 5.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.2 | 3.5 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 1.6 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 4.9 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.1 | 6.2 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.1 | 4.1 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 2.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 2.3 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.1 | 2.4 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 3.9 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 2.8 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.1 | 1.9 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 1.1 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 2.8 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.4 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.6 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.8 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 1.4 | 12.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 1.3 | 15.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.9 | 8.4 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.9 | 5.5 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.7 | 3.6 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.5 | 8.4 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.5 | 2.3 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.4 | 4.9 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.4 | 10.0 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.4 | 5.0 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.4 | 4.9 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.3 | 4.9 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.3 | 2.9 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.3 | 8.2 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.3 | 10.7 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.2 | 1.6 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.2 | 1.5 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.2 | 4.3 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.2 | 3.9 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.2 | 4.5 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.2 | 15.1 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.2 | 6.1 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.2 | 6.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.2 | 11.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.2 | 3.1 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 4.2 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 3.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 1.8 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 7.3 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 0.5 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 2.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 1.1 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 2.8 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 2.0 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 0.8 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 2.5 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 2.9 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 1.1 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.1 | 1.9 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.1 | 1.7 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 5.6 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.1 | 11.2 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 5.2 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 1.4 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.1 | 0.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 0.6 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 1.1 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 1.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.9 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.7 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 1.1 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 2.6 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 2.9 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 0.7 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 1.6 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.4 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.4 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 1.2 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.6 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |