Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
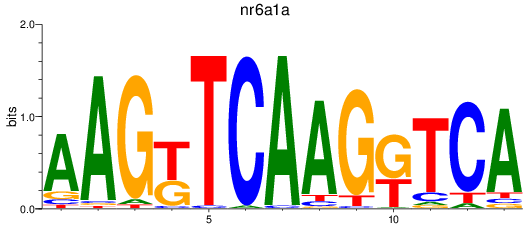
Results for nr6a1a
Z-value: 1.64
Transcription factors associated with nr6a1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr6a1a
|
ENSDARG00000101508 | nuclear receptor subfamily 6, group A, member 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr6a1a | dr11_v1_chr8_-_53044300_53044300 | -0.62 | 4.2e-03 | Click! |
Activity profile of nr6a1a motif
Sorted Z-values of nr6a1a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_22188117 | 9.29 |
ENSDART00000132890
|
crygm2d17
|
crystallin, gamma M2d17 |
| chr19_-_5135345 | 8.91 |
ENSDART00000151787
|
gnb3b
|
guanine nucleotide binding protein (G protein), beta polypeptide 3b |
| chr10_-_11383900 | 8.14 |
ENSDART00000101045
|
plac8.1
|
placenta-specific 8, tandem duplicate 1 |
| chr2_-_37210397 | 6.55 |
ENSDART00000084938
|
apoda.1
|
apolipoprotein Da, duplicate 1 |
| chr3_+_39540014 | 6.20 |
ENSDART00000074848
|
zgc:165423
|
zgc:165423 |
| chr3_+_13469704 | 6.05 |
ENSDART00000166806
|
zgc:77748
|
zgc:77748 |
| chr5_-_50992690 | 4.54 |
ENSDART00000149553
ENSDART00000097460 ENSDART00000192021 |
hmgcra
|
3-hydroxy-3-methylglutaryl-CoA reductase a |
| chr9_-_44939104 | 4.44 |
ENSDART00000192903
|
vil1
|
villin 1 |
| chr14_+_32838110 | 4.26 |
ENSDART00000158077
|
arr3b
|
arrestin 3b, retinal (X-arrestin) |
| chr12_+_34896956 | 4.10 |
ENSDART00000055415
|
prph2a
|
peripherin 2a (retinal degeneration, slow) |
| chr25_+_14092871 | 4.03 |
ENSDART00000067239
|
guca1g
|
guanylate cyclase activator 1g |
| chr14_-_33454595 | 4.01 |
ENSDART00000109615
ENSDART00000173267 ENSDART00000185737 ENSDART00000190989 |
tmem255a
|
transmembrane protein 255A |
| chr4_+_6643421 | 3.82 |
ENSDART00000099462
|
gpr85
|
G protein-coupled receptor 85 |
| chr25_+_33939728 | 3.77 |
ENSDART00000148537
|
roraa
|
RAR-related orphan receptor A, paralog a |
| chr19_-_703898 | 3.58 |
ENSDART00000181096
ENSDART00000121462 |
slc6a19a.2
|
solute carrier family 6 (neutral amino acid transporter), member 19a, tandem duplicate 2 |
| chr17_+_25414033 | 3.55 |
ENSDART00000001691
|
tdh2
|
L-threonine dehydrogenase 2 |
| chr8_+_44703864 | 3.53 |
ENSDART00000016225
|
star
|
steroidogenic acute regulatory protein |
| chr5_-_9090178 | 3.47 |
ENSDART00000091472
|
kcnv2b
|
potassium channel, subfamily V, member 2b |
| chr10_-_17284055 | 3.44 |
ENSDART00000167464
|
GNAZ
|
G protein subunit alpha z |
| chr25_+_3326885 | 3.33 |
ENSDART00000104866
|
ldhbb
|
lactate dehydrogenase Bb |
| chr1_-_50859053 | 3.29 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr17_+_18117358 | 3.20 |
ENSDART00000144894
|
bcl11ba
|
B cell CLL/lymphoma 11Ba |
| chr6_-_47246948 | 3.13 |
ENSDART00000162435
|
grm4
|
glutamate receptor, metabotropic 4 |
| chr6_+_41099787 | 3.08 |
ENSDART00000186884
|
fkbp5
|
FK506 binding protein 5 |
| chr12_+_46605600 | 3.05 |
ENSDART00000123533
|
FADS6
|
fatty acid desaturase 6 |
| chr13_-_2215213 | 3.05 |
ENSDART00000129773
|
mlip
|
muscular LMNA-interacting protein |
| chr10_-_16868211 | 2.98 |
ENSDART00000171755
|
stoml2
|
stomatin (EPB72)-like 2 |
| chr11_-_97817 | 2.95 |
ENSDART00000092903
|
elmo2
|
engulfment and cell motility 2 |
| chr14_+_32837914 | 2.89 |
ENSDART00000158888
|
arr3b
|
arrestin 3b, retinal (X-arrestin) |
| chr20_-_30370884 | 2.82 |
ENSDART00000062429
|
allc
|
allantoicase |
| chr4_-_17409533 | 2.82 |
ENSDART00000011943
|
pah
|
phenylalanine hydroxylase |
| chr20_-_43771871 | 2.78 |
ENSDART00000153304
|
matn3a
|
matrilin 3a |
| chr15_+_19682013 | 2.71 |
ENSDART00000127368
|
si:dkey-4p15.5
|
si:dkey-4p15.5 |
| chr6_+_42338309 | 2.69 |
ENSDART00000015277
|
gpx1b
|
glutathione peroxidase 1b |
| chr10_-_19801651 | 2.69 |
ENSDART00000080430
|
gfra2b
|
GDNF family receptor alpha 2b |
| chr3_+_19299309 | 2.63 |
ENSDART00000046297
ENSDART00000146955 |
ldlra
|
low density lipoprotein receptor a |
| chr6_+_48618512 | 2.56 |
ENSDART00000111190
|
FAM19A3
|
si:dkey-238f9.1 |
| chr25_+_7492663 | 2.53 |
ENSDART00000166496
|
cat
|
catalase |
| chr20_+_4392687 | 2.50 |
ENSDART00000187271
|
im:7142702
|
im:7142702 |
| chr10_-_42685512 | 2.50 |
ENSDART00000081347
|
stc1l
|
stanniocalcin 1, like |
| chr11_+_45287541 | 2.45 |
ENSDART00000165321
ENSDART00000173116 |
pycr1b
|
pyrroline-5-carboxylate reductase 1b |
| chr4_+_7827261 | 2.39 |
ENSDART00000129568
|
phyh
|
phytanoyl-CoA 2-hydroxylase |
| chr7_-_20731078 | 2.37 |
ENSDART00000188267
|
chd3
|
chromodomain helicase DNA binding protein 3 |
| chr24_-_17047918 | 2.37 |
ENSDART00000020204
|
msrb2
|
methionine sulfoxide reductase B2 |
| chr21_-_40174647 | 2.35 |
ENSDART00000183738
ENSDART00000076840 ENSDART00000145109 |
slco2b1
|
solute carrier organic anion transporter family, member 2B1 |
| chr23_-_35756115 | 2.35 |
ENSDART00000043429
|
jph2
|
junctophilin 2 |
| chr25_+_3327071 | 2.35 |
ENSDART00000136131
ENSDART00000133243 |
ldhbb
|
lactate dehydrogenase Bb |
| chr13_-_30662403 | 2.32 |
ENSDART00000012457
|
si:dkey-275b16.2
|
si:dkey-275b16.2 |
| chr13_-_21688176 | 2.32 |
ENSDART00000063825
|
sprn
|
shadow of prion protein |
| chr16_+_24626448 | 2.28 |
ENSDART00000153591
|
si:dkey-56f14.7
|
si:dkey-56f14.7 |
| chr10_-_24371312 | 2.23 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr5_+_9428876 | 2.19 |
ENSDART00000081791
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr19_-_103289 | 2.19 |
ENSDART00000143118
|
adgrb1b
|
adhesion G protein-coupled receptor B1b |
| chr7_+_19482084 | 2.18 |
ENSDART00000173873
|
si:ch211-212k18.7
|
si:ch211-212k18.7 |
| chr12_+_45676667 | 2.18 |
ENSDART00000016553
|
si:ch73-111m19.2
|
si:ch73-111m19.2 |
| chr6_+_39836474 | 2.16 |
ENSDART00000112637
|
smarcc2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 |
| chr24_-_17023392 | 2.14 |
ENSDART00000106058
|
ptgdsb.2
|
prostaglandin D2 synthase b, tandem duplicate 2 |
| chr9_+_31795343 | 2.14 |
ENSDART00000139584
|
itgbl1
|
integrin, beta-like 1 |
| chr6_-_56111829 | 2.12 |
ENSDART00000154397
|
gcnt7
|
glucosaminyl (N-acetyl) transferase family member 7 |
| chr21_-_30658509 | 2.11 |
ENSDART00000139764
|
si:dkey-22f5.9
|
si:dkey-22f5.9 |
| chr6_+_39506043 | 2.10 |
ENSDART00000086260
|
CR388364.1
|
|
| chr16_+_14033121 | 2.06 |
ENSDART00000135844
|
rusc1
|
RUN and SH3 domain containing 1 |
| chr1_-_45553602 | 2.05 |
ENSDART00000143664
|
grin2bb
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2B, genome duplicate b |
| chr2_+_37110504 | 2.04 |
ENSDART00000132794
ENSDART00000042974 |
slc1a8b
|
solute carrier family 1 (glutamate transporter), member 8b |
| chr8_+_19356072 | 2.03 |
ENSDART00000063272
|
mpeg1.2
|
macrophage expressed 1, tandem duplicate 2 |
| chr10_-_39321367 | 2.01 |
ENSDART00000129647
|
smtlb
|
somatolactin beta |
| chr16_+_10776688 | 2.01 |
ENSDART00000161969
ENSDART00000172657 |
atp1a3b
|
ATPase Na+/K+ transporting subunit alpha 3b |
| chr5_+_6892195 | 2.01 |
ENSDART00000048201
|
gb:cr929477
|
expressed sequence CR929477 |
| chr17_+_18117029 | 2.00 |
ENSDART00000154646
ENSDART00000179739 |
bcl11ba
|
B cell CLL/lymphoma 11Ba |
| chr2_+_52232630 | 1.97 |
ENSDART00000006216
|
plpp2a
|
phospholipid phosphatase 2a |
| chr12_+_33395748 | 1.95 |
ENSDART00000129458
|
fasn
|
fatty acid synthase |
| chr1_-_46632948 | 1.93 |
ENSDART00000148893
ENSDART00000053232 |
cdadc1
|
cytidine and dCMP deaminase domain containing 1 |
| chr8_+_3820134 | 1.92 |
ENSDART00000122454
|
citb
|
citron rho-interacting serine/threonine kinase b |
| chr16_-_12319822 | 1.92 |
ENSDART00000127453
ENSDART00000184526 |
trpv6
|
transient receptor potential cation channel, subfamily V, member 6 |
| chr16_+_47428721 | 1.89 |
ENSDART00000180597
ENSDART00000032309 |
slc25a32b
|
solute carrier family 25 (mitochondrial folate carrier), member 32b |
| chr12_-_4346085 | 1.87 |
ENSDART00000112433
|
ca15c
|
carbonic anhydrase XV c |
| chr2_-_24289641 | 1.86 |
ENSDART00000128784
ENSDART00000123565 ENSDART00000141922 ENSDART00000184550 ENSDART00000191469 |
myh7l
|
myosin heavy chain 7-like |
| chr8_+_19514294 | 1.84 |
ENSDART00000170622
|
si:ch73-281k2.5
|
si:ch73-281k2.5 |
| chr25_-_13188678 | 1.84 |
ENSDART00000125754
|
si:ch211-147m6.1
|
si:ch211-147m6.1 |
| chr15_-_28247583 | 1.83 |
ENSDART00000112967
|
rilp
|
Rab interacting lysosomal protein |
| chr15_+_26600611 | 1.82 |
ENSDART00000155352
|
slc47a3
|
solute carrier family 47 (multidrug and toxin extrusion), member 3 |
| chr5_+_9434288 | 1.82 |
ENSDART00000162089
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr16_-_17162485 | 1.82 |
ENSDART00000123011
|
iffo1b
|
intermediate filament family orphan 1b |
| chr19_-_5103313 | 1.76 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr9_-_21238616 | 1.75 |
ENSDART00000191840
ENSDART00000189127 |
cox17
|
COX17 cytochrome c oxidase copper chaperone |
| chr4_-_4932619 | 1.74 |
ENSDART00000103293
|
ndufa5
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5 |
| chr24_-_28245872 | 1.74 |
ENSDART00000167861
|
cox4i1l
|
cytochrome c oxidase subunit IV isoform 1, like |
| chr9_+_13682133 | 1.72 |
ENSDART00000175639
|
mpp4a
|
membrane protein, palmitoylated 4a (MAGUK p55 subfamily member 4) |
| chr15_+_19681718 | 1.70 |
ENSDART00000164803
|
si:dkey-4p15.5
|
si:dkey-4p15.5 |
| chr5_-_37103487 | 1.69 |
ENSDART00000149211
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr11_-_18799827 | 1.69 |
ENSDART00000185438
ENSDART00000189116 ENSDART00000180504 |
pfkfb2b
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2b |
| chr7_-_16205471 | 1.68 |
ENSDART00000173584
|
btr05
|
bloodthirsty-related gene family, member 5 |
| chr18_+_8340886 | 1.68 |
ENSDART00000081132
|
cpt1b
|
carnitine palmitoyltransferase 1B (muscle) |
| chr8_-_29713595 | 1.67 |
ENSDART00000131988
ENSDART00000077637 |
mpeg1.1
|
macrophage expressed 1, tandem duplicate 1 |
| chr16_-_18960613 | 1.66 |
ENSDART00000183197
|
fhod3b
|
formin homology 2 domain containing 3b |
| chr9_-_21238159 | 1.66 |
ENSDART00000146764
ENSDART00000102143 |
cox17
|
COX17 cytochrome c oxidase copper chaperone |
| chr23_-_19953089 | 1.65 |
ENSDART00000153828
|
atp2b3b
|
ATPase plasma membrane Ca2+ transporting 3b |
| chr1_-_55785722 | 1.65 |
ENSDART00000142069
ENSDART00000043933 |
ndufb7
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 7 |
| chr10_-_42131408 | 1.64 |
ENSDART00000076693
|
stambpa
|
STAM binding protein a |
| chr19_-_5103141 | 1.64 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr15_-_14375452 | 1.61 |
ENSDART00000160675
ENSDART00000164028 ENSDART00000171642 |
dpf1
|
D4, zinc and double PHD fingers family 1 |
| chr1_-_22726233 | 1.61 |
ENSDART00000140920
|
prom1b
|
prominin 1 b |
| chr11_-_7320211 | 1.60 |
ENSDART00000091664
|
apc2
|
adenomatosis polyposis coli 2 |
| chr25_-_34512102 | 1.56 |
ENSDART00000087450
|
klf13
|
Kruppel-like factor 13 |
| chr24_-_20808283 | 1.54 |
ENSDART00000143759
|
vipr1b
|
vasoactive intestinal peptide receptor 1b |
| chr17_-_39880725 | 1.53 |
ENSDART00000184030
|
zmp:0000000545
|
zmp:0000000545 |
| chr22_-_5518117 | 1.50 |
ENSDART00000164613
|
CABZ01064972.1
|
|
| chr21_-_22737228 | 1.49 |
ENSDART00000151366
|
fbxo40.2
|
F-box protein 40, tandem duplicate 2 |
| chr8_+_2878756 | 1.49 |
ENSDART00000168107
|
crb2b
|
crumbs family member 2b |
| chr23_+_16633951 | 1.47 |
ENSDART00000109537
ENSDART00000193323 |
snphb
|
syntaphilin b |
| chr6_+_52235441 | 1.46 |
ENSDART00000056319
|
cox6c
|
cytochrome c oxidase subunit VIc |
| chr5_+_42124706 | 1.43 |
ENSDART00000020044
ENSDART00000156372 |
shpk
|
sedoheptulokinase |
| chr14_-_36412473 | 1.42 |
ENSDART00000128244
ENSDART00000138376 |
asb5a
|
ankyrin repeat and SOCS box containing 5a |
| chr9_-_48700806 | 1.42 |
ENSDART00000026210
|
rdh1
|
retinol dehydrogenase 1 |
| chr20_-_14665002 | 1.41 |
ENSDART00000152816
|
scrn2
|
secernin 2 |
| chr1_+_34243650 | 1.40 |
ENSDART00000147201
|
slc5a7a
|
solute carrier family 5 (sodium/choline cotransporter), member 7a |
| chr11_-_27874116 | 1.40 |
ENSDART00000180579
|
eif4g3a
|
eukaryotic translation initiation factor 4 gamma, 3a |
| chr14_-_3174570 | 1.39 |
ENSDART00000163585
|
csf1ra
|
colony stimulating factor 1 receptor, a |
| chr9_-_54001502 | 1.38 |
ENSDART00000085253
|
mid1
|
midline 1 |
| chr19_-_42571829 | 1.37 |
ENSDART00000102606
|
zgc:103438
|
zgc:103438 |
| chr3_+_4346854 | 1.37 |
ENSDART00000004273
|
si:dkey-73p2.3
|
si:dkey-73p2.3 |
| chr25_-_16742438 | 1.36 |
ENSDART00000156091
|
galnt8a.1
|
polypeptide N-acetylgalactosaminyltransferase 8a, tandem duplicate 1 |
| chr7_+_26138240 | 1.36 |
ENSDART00000193750
ENSDART00000184942 |
nat16
|
N-acetyltransferase 16 |
| chr13_-_3351708 | 1.32 |
ENSDART00000042875
|
si:ch73-193i2.2
|
si:ch73-193i2.2 |
| chr2_+_7818368 | 1.32 |
ENSDART00000007068
|
kcnmb2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr15_-_46718759 | 1.31 |
ENSDART00000085926
ENSDART00000154577 |
zgc:162872
|
zgc:162872 |
| chr19_-_27858033 | 1.31 |
ENSDART00000103898
ENSDART00000144884 |
srd5a1
|
steroid-5-alpha-reductase, alpha polypeptide 1 (3-oxo-5 alpha-steroid delta 4-dehydrogenase alpha 1) |
| chr5_-_38064065 | 1.30 |
ENSDART00000137181
|
si:dkey-111e8.5
|
si:dkey-111e8.5 |
| chr9_-_46276626 | 1.30 |
ENSDART00000165238
|
hdac4
|
histone deacetylase 4 |
| chr7_+_17096281 | 1.30 |
ENSDART00000035558
|
htatip2
|
HIV-1 Tat interactive protein 2 |
| chr3_-_30609659 | 1.29 |
ENSDART00000182516
ENSDART00000187047 ENSDART00000110597 |
syt3
|
synaptotagmin III |
| chr23_+_28092083 | 1.29 |
ENSDART00000053958
|
c1galt1a
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1a |
| chr17_+_33311784 | 1.28 |
ENSDART00000156229
|
si:ch211-132f19.7
|
si:ch211-132f19.7 |
| chr7_+_16033923 | 1.28 |
ENSDART00000161669
ENSDART00000114062 |
immp1l
|
inner mitochondrial membrane peptidase subunit 1 |
| chr11_+_27133560 | 1.28 |
ENSDART00000158411
|
hdac11
|
histone deacetylase 11 |
| chr5_-_26936861 | 1.27 |
ENSDART00000191032
|
htra4
|
HtrA serine peptidase 4 |
| chr1_+_50085640 | 1.27 |
ENSDART00000158988
ENSDART00000165973 |
npnt
|
nephronectin |
| chr7_+_65261354 | 1.24 |
ENSDART00000168388
|
bco1
|
beta-carotene oxygenase 1 |
| chr24_-_25574967 | 1.22 |
ENSDART00000189828
|
cnksr2a
|
connector enhancer of kinase suppressor of Ras 2a |
| chr20_-_23219964 | 1.21 |
ENSDART00000144933
|
dcun1d4
|
DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae) |
| chr3_-_58455289 | 1.20 |
ENSDART00000052179
|
cdr2a
|
cerebellar degeneration-related protein 2a |
| chr21_+_15713097 | 1.19 |
ENSDART00000015841
|
gstt1b
|
glutathione S-transferase theta 1b |
| chr1_+_50085440 | 1.19 |
ENSDART00000018469
ENSDART00000134988 |
npnt
|
nephronectin |
| chr3_+_1749793 | 1.19 |
ENSDART00000149308
|
si:dkeyp-52c3.7
|
si:dkeyp-52c3.7 |
| chr25_-_35963158 | 1.17 |
ENSDART00000153612
|
snx20
|
sorting nexin 20 |
| chr21_-_43040780 | 1.17 |
ENSDART00000187037
|
jakmip2
|
janus kinase and microtubule interacting protein 2 |
| chr17_-_36904838 | 1.17 |
ENSDART00000188030
|
BX571720.1
|
|
| chr22_-_5933844 | 1.17 |
ENSDART00000163370
ENSDART00000189331 |
si:rp71-36a1.2
|
si:rp71-36a1.2 |
| chr2_+_24868010 | 1.16 |
ENSDART00000078838
|
rab3aa
|
RAB3A, member RAS oncogene family, a |
| chr10_-_25543227 | 1.11 |
ENSDART00000007778
|
grik1a
|
glutamate receptor, ionotropic, kainate 1a |
| chr16_+_50741154 | 1.11 |
ENSDART00000101627
|
IGLON5
|
zgc:110372 |
| chr5_-_41834999 | 1.10 |
ENSDART00000135772
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr2_+_38924975 | 1.10 |
ENSDART00000109219
|
rem2
|
RAS (RAD and GEM)-like GTP binding 2 |
| chr20_+_9781932 | 1.09 |
ENSDART00000053841
|
ddhd1b
|
DDHD domain containing 1b |
| chr2_+_51028269 | 1.08 |
ENSDART00000161254
|
eef1da
|
eukaryotic translation elongation factor 1 delta a (guanine nucleotide exchange protein) |
| chr13_+_31192503 | 1.07 |
ENSDART00000147936
|
ptpn20
|
protein tyrosine phosphatase, non-receptor type 20 |
| chr4_-_34776147 | 1.07 |
ENSDART00000167498
|
si:dkey-146m20.14
|
si:dkey-146m20.14 |
| chr1_-_53476758 | 1.06 |
ENSDART00000074221
|
b3gnt2a
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2a |
| chr5_+_23622177 | 1.05 |
ENSDART00000121504
|
cx27.5
|
connexin 27.5 |
| chr18_-_226800 | 1.04 |
ENSDART00000165180
|
tarsl2
|
threonyl-tRNA synthetase-like 2 |
| chr5_-_69004007 | 1.04 |
ENSDART00000137443
|
ank1a
|
ankyrin 1, erythrocytic a |
| chr13_-_1085961 | 1.02 |
ENSDART00000171872
ENSDART00000166598 |
plek
|
pleckstrin |
| chr10_+_25219728 | 1.02 |
ENSDART00000193829
|
grm5a
|
glutamate receptor, metabotropic 5a |
| chr7_+_65261576 | 1.00 |
ENSDART00000169566
|
bco1
|
beta-carotene oxygenase 1 |
| chr7_-_56606752 | 1.00 |
ENSDART00000138714
|
sult5a1
|
sulfotransferase family 5A, member 1 |
| chr24_-_25256230 | 0.99 |
ENSDART00000155780
|
hhla2b.1
|
HERV-H LTR-associating 2b, tandem duplicate 1 |
| chr17_-_3190070 | 0.99 |
ENSDART00000115027
|
tmem151bb
|
transmembrane protein 151Bb |
| chr4_-_63166758 | 0.99 |
ENSDART00000192765
|
CR450780.1
|
|
| chr24_-_17029374 | 0.99 |
ENSDART00000039267
|
ptgdsb.1
|
prostaglandin D2 synthase b, tandem duplicate 1 |
| chr7_-_58130703 | 0.97 |
ENSDART00000172082
|
ank2b
|
ankyrin 2b, neuronal |
| chr19_+_32855139 | 0.97 |
ENSDART00000052082
|
rpl30
|
ribosomal protein L30 |
| chr19_-_32487469 | 0.97 |
ENSDART00000050130
|
gmpr
|
guanosine monophosphate reductase |
| chr9_-_14055959 | 0.96 |
ENSDART00000146675
|
fer1l6
|
fer-1-like family member 6 |
| chr15_+_46386261 | 0.96 |
ENSDART00000191793
|
igsf11
|
immunoglobulin superfamily member 11 |
| chr8_-_18316035 | 0.96 |
ENSDART00000134534
|
rnf220b
|
ring finger protein 220b |
| chr9_-_48701035 | 0.94 |
ENSDART00000178332
|
rdh1
|
retinol dehydrogenase 1 |
| chr21_-_7687544 | 0.94 |
ENSDART00000134519
|
pde8b
|
phosphodiesterase 8B |
| chr1_-_37000507 | 0.93 |
ENSDART00000160637
|
nr3c2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr15_+_22722684 | 0.93 |
ENSDART00000156760
|
grik4
|
glutamate receptor, ionotropic, kainate 4 |
| chr18_+_8912113 | 0.93 |
ENSDART00000147467
|
tmem243a
|
transmembrane protein 243, mitochondrial a |
| chr14_+_41518257 | 0.93 |
ENSDART00000050037
|
chrnb3b
|
cholinergic receptor, nicotinic, beta 3b (neuronal) |
| chr1_-_22652424 | 0.93 |
ENSDART00000036797
|
uchl1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
| chr4_-_70263609 | 0.92 |
ENSDART00000163868
|
si:dkey-3h2.3
|
si:dkey-3h2.3 |
| chr8_-_31393429 | 0.92 |
ENSDART00000192369
|
nim1k
|
NIM1 serine/threonine protein kinase |
| chr19_-_41371978 | 0.91 |
ENSDART00000166063
ENSDART00000170343 |
slc25a13
|
solute carrier family 25 (aspartate/glutamate carrier), member 13 |
| chr18_-_21806613 | 0.91 |
ENSDART00000145721
|
nrn1la
|
neuritin 1-like a |
| chr4_-_71267777 | 0.91 |
ENSDART00000169664
|
si:dkeyp-51g9.4
|
si:dkeyp-51g9.4 |
| chr4_-_63558838 | 0.89 |
ENSDART00000191708
ENSDART00000190740 ENSDART00000186476 |
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr3_-_584950 | 0.89 |
ENSDART00000164752
|
dicp1.1
|
diverse immunoglobulin domain-containing protein 1.1 |
| chr25_-_16742636 | 0.88 |
ENSDART00000192486
ENSDART00000192711 |
galnt8a.1
|
polypeptide N-acetylgalactosaminyltransferase 8a, tandem duplicate 1 |
| chr4_+_6572364 | 0.87 |
ENSDART00000122574
|
ppp1r3aa
|
protein phosphatase 1, regulatory subunit 3Aa |
| chr10_-_41450367 | 0.87 |
ENSDART00000122682
ENSDART00000189549 |
cabp1b
|
calcium binding protein 1b |
| chr6_-_30932078 | 0.87 |
ENSDART00000028612
|
pde4ba
|
phosphodiesterase 4B, cAMP-specific a |
| chr1_+_17527342 | 0.86 |
ENSDART00000139702
ENSDART00000140076 ENSDART00000005593 |
casp3a
|
caspase 3, apoptosis-related cysteine peptidase a |
| chr22_-_23781083 | 0.86 |
ENSDART00000166563
ENSDART00000170458 ENSDART00000166158 ENSDART00000171246 |
cfhl3
|
complement factor H like 3 |
| chr22_-_17462895 | 0.85 |
ENSDART00000160415
|
si:ch211-197g15.6
|
si:ch211-197g15.6 |
| chr22_-_17781213 | 0.85 |
ENSDART00000137984
|
si:ch73-63e15.2
|
si:ch73-63e15.2 |
| chr22_-_6229275 | 0.84 |
ENSDART00000146045
ENSDART00000179730 |
si:ch211-274k16.2
|
si:ch211-274k16.2 |
| chr16_-_6391275 | 0.83 |
ENSDART00000060550
|
ndufb9
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 9 |
| chr5_+_23624684 | 0.82 |
ENSDART00000051539
|
cx27.5
|
connexin 27.5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr6a1a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.4 | GO:2000392 | lamellipodium morphogenesis(GO:0072673) regulation of lamellipodium morphogenesis(GO:2000392) |
| 1.0 | 6.2 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.9 | 2.8 | GO:0000256 | allantoin catabolic process(GO:0000256) |
| 0.9 | 3.4 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.6 | 7.2 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.6 | 2.4 | GO:0030091 | protein repair(GO:0030091) |
| 0.5 | 2.2 | GO:0090387 | phagosome maturation involved in apoptotic cell clearance(GO:0090386) phagolysosome assembly involved in apoptotic cell clearance(GO:0090387) |
| 0.5 | 1.9 | GO:0070316 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.5 | 1.4 | GO:1900620 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.4 | 2.7 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.4 | 1.2 | GO:2000434 | regulation of protein neddylation(GO:2000434) |
| 0.4 | 3.6 | GO:0006566 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.4 | 1.2 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.3 | 1.4 | GO:0071674 | regulation of macrophage chemotaxis(GO:0010758) positive regulation of macrophage chemotaxis(GO:0010759) regulation of odontogenesis(GO:0042481) mononuclear cell migration(GO:0071674) regulation of mononuclear cell migration(GO:0071675) |
| 0.3 | 2.4 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.3 | 1.0 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.3 | 3.0 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.3 | 5.9 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.3 | 1.3 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.3 | 1.3 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.3 | 1.5 | GO:0061438 | renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.3 | 1.5 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.3 | 5.1 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.2 | 0.7 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.2 | 2.8 | GO:1902222 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.2 | 0.9 | GO:0071387 | cellular response to cortisol stimulus(GO:0071387) |
| 0.2 | 1.3 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.2 | 1.1 | GO:0019860 | uracil catabolic process(GO:0006212) beta-alanine metabolic process(GO:0019482) beta-alanine biosynthetic process(GO:0019483) uracil metabolic process(GO:0019860) |
| 0.2 | 6.5 | GO:0007568 | aging(GO:0007568) |
| 0.2 | 1.0 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.2 | 3.6 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.2 | 2.0 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.2 | 1.3 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.2 | 2.4 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 0.4 | GO:0033212 | iron assimilation(GO:0033212) |
| 0.1 | 1.6 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.1 | 1.6 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.4 | GO:0035376 | sterol import(GO:0035376) recognition of apoptotic cell(GO:0043654) cholesterol import(GO:0070508) |
| 0.1 | 0.4 | GO:0006678 | glucosylceramide metabolic process(GO:0006678) |
| 0.1 | 0.4 | GO:0097237 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.1 | 2.5 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 2.0 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.1 | 2.0 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 1.7 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 1.5 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.1 | 3.1 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 | 0.4 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.1 | 0.7 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.1 | 0.7 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 0.9 | GO:0015813 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.4 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.1 | 1.8 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 1.0 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.1 | 2.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 1.1 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 1.3 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 0.4 | GO:0035889 | otolith tethering(GO:0035889) |
| 0.1 | 0.3 | GO:0070255 | regulation of mucus secretion(GO:0070255) positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 0.6 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 9.3 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.1 | 1.0 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.1 | 2.1 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.1 | 1.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 3.8 | GO:0051592 | response to calcium ion(GO:0051592) |
| 0.1 | 0.4 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) glycosylceramide biosynthetic process(GO:0046476) |
| 0.1 | 2.4 | GO:0021587 | cerebellum morphogenesis(GO:0021587) |
| 0.1 | 1.7 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.1 | 1.4 | GO:0071219 | cellular response to molecule of bacterial origin(GO:0071219) cellular response to lipopolysaccharide(GO:0071222) |
| 0.1 | 1.6 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 0.9 | GO:0070672 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.1 | 0.3 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 1.0 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.0 | 0.9 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 1.1 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.9 | GO:0005979 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.0 | 1.2 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 1.0 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 2.2 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.6 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.5 | GO:0070897 | DNA-templated transcriptional preinitiation complex assembly(GO:0070897) |
| 0.0 | 2.5 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 1.7 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 1.3 | GO:0046058 | cAMP metabolic process(GO:0046058) |
| 0.0 | 0.3 | GO:0043407 | negative regulation of MAP kinase activity(GO:0043407) |
| 0.0 | 1.2 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 1.9 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 1.5 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 3.0 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.0 | 14.9 | GO:0045944 | positive regulation of transcription from RNA polymerase II promoter(GO:0045944) |
| 0.0 | 0.7 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.3 | GO:0034112 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.0 | 0.3 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.0 | 0.9 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.6 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.0 | 1.1 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.1 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.0 | 0.1 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.7 | GO:0007548 | sex differentiation(GO:0007548) |
| 0.0 | 0.6 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.5 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 1.0 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 1.5 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 1.2 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 1.7 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.0 | 0.1 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.1 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 2.2 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 3.2 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 14.0 | GO:0007186 | G-protein coupled receptor signaling pathway(GO:0007186) |
| 0.0 | 0.7 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.5 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.0 | 0.3 | GO:0050727 | regulation of inflammatory response(GO:0050727) |
| 0.0 | 0.9 | GO:0051480 | regulation of cytosolic calcium ion concentration(GO:0051480) |
| 0.0 | 0.3 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.7 | GO:0007254 | JNK cascade(GO:0007254) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.4 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.5 | 2.3 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.3 | 1.0 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.3 | 1.3 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.2 | 0.9 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.2 | 1.6 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.2 | 1.6 | GO:0071914 | prominosome(GO:0071914) |
| 0.2 | 7.4 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.2 | 1.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 2.0 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 8.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 0.6 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.1 | 1.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 1.6 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 4.5 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 2.3 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 3.0 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 2.9 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 0.8 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 1.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.5 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.1 | 1.0 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 3.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 4.6 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 0.9 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 2.1 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.7 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 2.5 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.8 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 1.4 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 4.2 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.7 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 1.0 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.9 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 1.3 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 2.6 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 3.4 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 1.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 12.9 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.4 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.8 | 5.7 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.8 | 3.1 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.8 | 3.0 | GO:0005521 | lamin binding(GO:0005521) |
| 0.7 | 2.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.7 | 3.6 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.7 | 3.4 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.7 | 2.0 | GO:0070186 | growth hormone receptor binding(GO:0005131) growth hormone activity(GO:0070186) |
| 0.7 | 2.6 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.6 | 1.9 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.6 | 8.9 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.6 | 2.4 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.5 | 4.0 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.5 | 2.5 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.5 | 2.3 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.4 | 1.4 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.3 | 2.4 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.3 | 1.3 | GO:0033765 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) enone reductase activity(GO:0035671) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.3 | 1.0 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.3 | 1.3 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.3 | 1.9 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.3 | 2.8 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.3 | 2.8 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.3 | 1.4 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.3 | 2.5 | GO:0004096 | catalase activity(GO:0004096) |
| 0.3 | 1.5 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.2 | 2.2 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.2 | 0.9 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.2 | 1.1 | GO:0002058 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.2 | 2.0 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.2 | 2.4 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.2 | 4.1 | GO:0098988 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.2 | 3.8 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.2 | 0.8 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.1 | 0.4 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.1 | 0.6 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.1 | 1.0 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 1.7 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 1.8 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.1 | 9.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 3.1 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.5 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.1 | 0.7 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 2.0 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 5.4 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 0.5 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.1 | 0.9 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.1 | 1.3 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.7 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 0.4 | GO:0071814 | low-density lipoprotein particle binding(GO:0030169) lipoprotein particle binding(GO:0071813) protein-lipid complex binding(GO:0071814) |
| 0.1 | 0.6 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.7 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 4.4 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 2.7 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 1.1 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 1.3 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 4.2 | GO:0050661 | NADP binding(GO:0050661) |
| 0.1 | 1.7 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 1.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.9 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 1.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 0.5 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.1 | 1.8 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 1.8 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.7 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.5 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 0.6 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.1 | 4.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 1.9 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.5 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 0.3 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 0.6 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.1 | 1.7 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 0.9 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 1.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 1.4 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.1 | 0.6 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.1 | 1.0 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 3.0 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 1.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 1.3 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.4 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.4 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.9 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 1.2 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.4 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 1.1 | GO:0004970 | ionotropic glutamate receptor activity(GO:0004970) |
| 0.0 | 0.8 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.9 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 2.6 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 2.1 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 1.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 1.3 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 3.8 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 7.2 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 0.7 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 1.9 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.1 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.0 | 6.9 | GO:0008236 | serine-type peptidase activity(GO:0008236) serine hydrolase activity(GO:0017171) |
| 0.0 | 0.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0004960 | thromboxane receptor activity(GO:0004960) |
| 0.0 | 0.1 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 4.4 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.3 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.5 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 3.3 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 1.4 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 0.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 2.2 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.3 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 3.7 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 1.8 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 0.3 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 1.0 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.4 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 4.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.0 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 1.9 | GO:0005262 | calcium channel activity(GO:0005262) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.4 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.2 | 1.7 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 1.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 1.4 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 0.7 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 3.0 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 2.1 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 2.5 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 3.1 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 1.3 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 2.5 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 2.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.6 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.9 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.9 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.5 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.5 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.4 | 3.1 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.3 | 2.4 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.2 | 4.8 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 3.4 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 2.0 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.1 | 1.7 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 2.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 6.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.0 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 4.3 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 1.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 0.7 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.1 | 0.7 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 0.9 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.0 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 0.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 2.2 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.1 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.6 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.3 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.9 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.7 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 1.6 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |