Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
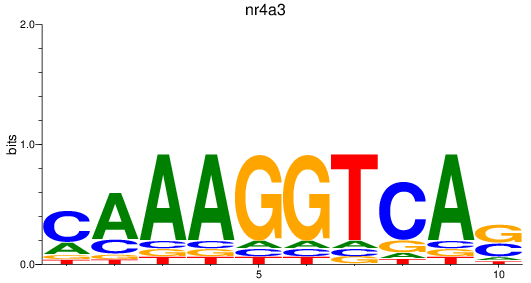
Results for nr4a3
Z-value: 0.62
Transcription factors associated with nr4a3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr4a3
|
ENSDARG00000055854 | nuclear receptor subfamily 4, group A, member 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr4a3 | dr11_v1_chr16_+_27345383_27345383 | -0.76 | 1.5e-04 | Click! |
Activity profile of nr4a3 motif
Sorted Z-values of nr4a3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_16451375 | 3.77 |
ENSDART00000192700
ENSDART00000128835 |
wu:fc23c09
|
wu:fc23c09 |
| chr16_-_45910050 | 2.71 |
ENSDART00000133213
|
afp4
|
antifreeze protein type IV |
| chr22_+_14102397 | 2.12 |
ENSDART00000146397
ENSDART00000137530 |
he1.3
|
hatching enzyme 1, tandem duplicate 3 |
| chr13_-_37127970 | 1.96 |
ENSDART00000135510
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr23_+_32335871 | 1.91 |
ENSDART00000149698
|
slc39a5
|
solute carrier family 39 (zinc transporter), member 5 |
| chr8_-_40183197 | 1.79 |
ENSDART00000005118
|
gpx8
|
glutathione peroxidase 8 (putative) |
| chr23_+_42396934 | 1.73 |
ENSDART00000169821
|
cyp2aa8
|
cytochrome P450, family 2, subfamily AA, polypeptide 8 |
| chr19_-_35439237 | 1.69 |
ENSDART00000145883
|
anln
|
anillin, actin binding protein |
| chr5_-_14509137 | 1.53 |
ENSDART00000180742
|
si:ch211-244o22.2
|
si:ch211-244o22.2 |
| chr4_+_15942075 | 1.40 |
ENSDART00000147964
|
si:dkey-117n7.2
|
si:dkey-117n7.2 |
| chr22_-_14262115 | 1.34 |
ENSDART00000168264
|
si:ch211-246m6.5
|
si:ch211-246m6.5 |
| chr21_+_8239544 | 1.23 |
ENSDART00000122773
|
nr6a1b
|
nuclear receptor subfamily 6, group A, member 1b |
| chr20_+_36806398 | 1.22 |
ENSDART00000153317
|
abracl
|
ABRA C-terminal like |
| chr8_+_39634114 | 1.20 |
ENSDART00000144293
|
msi1
|
musashi RNA-binding protein 1 |
| chr1_-_39859626 | 1.20 |
ENSDART00000053763
|
dctd
|
dCMP deaminase |
| chr10_+_10351685 | 1.19 |
ENSDART00000109432
|
cercam
|
cerebral endothelial cell adhesion molecule |
| chr24_-_12770357 | 1.16 |
ENSDART00000060826
|
ipo4
|
importin 4 |
| chr22_-_26100282 | 1.09 |
ENSDART00000166075
|
pdgfaa
|
platelet-derived growth factor alpha polypeptide a |
| chr22_-_17595310 | 1.07 |
ENSDART00000099056
|
gpx4a
|
glutathione peroxidase 4a |
| chr17_-_28097760 | 1.06 |
ENSDART00000149861
|
kdm1a
|
lysine (K)-specific demethylase 1a |
| chr22_-_7050 | 1.04 |
ENSDART00000127829
|
atad3
|
ATPase family, AAA domain containing 3 |
| chr6_+_7414215 | 1.03 |
ENSDART00000049339
|
sox21a
|
SRY (sex determining region Y)-box 21a |
| chr22_-_26005894 | 1.03 |
ENSDART00000105088
|
pdgfaa
|
platelet-derived growth factor alpha polypeptide a |
| chr21_-_25250594 | 1.01 |
ENSDART00000163862
|
nfrkb
|
nuclear factor related to kappaB binding protein |
| chr7_+_23943597 | 1.01 |
ENSDART00000157408
|
si:dkey-183c6.7
|
si:dkey-183c6.7 |
| chr1_+_39859782 | 1.00 |
ENSDART00000149984
|
irf2a
|
interferon regulatory factor 2a |
| chr1_+_24557414 | 0.98 |
ENSDART00000076519
|
dctpp1
|
dCTP pyrophosphatase 1 |
| chr5_+_36896933 | 0.97 |
ENSDART00000151984
|
srsf7a
|
serine/arginine-rich splicing factor 7a |
| chr3_-_49925313 | 0.93 |
ENSDART00000164361
|
gcgra
|
glucagon receptor a |
| chr25_+_418932 | 0.92 |
ENSDART00000059193
|
prtgb
|
protogenin homolog b (Gallus gallus) |
| chr23_-_16737161 | 0.91 |
ENSDART00000132573
|
si:ch211-224l10.4
|
si:ch211-224l10.4 |
| chr9_+_500052 | 0.89 |
ENSDART00000166707
|
CU984600.1
|
|
| chr4_-_9191220 | 0.87 |
ENSDART00000156919
|
hcfc2
|
host cell factor C2 |
| chr8_-_22542467 | 0.82 |
ENSDART00000182588
ENSDART00000134542 |
csde1
|
cold shock domain containing E1, RNA-binding |
| chr25_-_35095129 | 0.81 |
ENSDART00000099866
ENSDART00000099868 |
kif15
|
kinesin family member 15 |
| chr24_-_28419444 | 0.81 |
ENSDART00000105749
|
nrros
|
negative regulator of reactive oxygen species |
| chr19_-_1947403 | 0.81 |
ENSDART00000113951
ENSDART00000151293 ENSDART00000134074 |
znrf2a
|
zinc and ring finger 2a |
| chr1_-_57172294 | 0.80 |
ENSDART00000063774
|
rac1l
|
Rac family small GTPase 1, like |
| chr2_-_17115256 | 0.78 |
ENSDART00000190488
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr1_+_38153944 | 0.77 |
ENSDART00000135666
|
galnt7
|
UDP-N-acetyl-alpha-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase 7 |
| chr17_+_48314724 | 0.76 |
ENSDART00000125617
|
smoc1
|
SPARC related modular calcium binding 1 |
| chr19_-_17972734 | 0.76 |
ENSDART00000126298
|
ints8
|
integrator complex subunit 8 |
| chr8_-_21110233 | 0.74 |
ENSDART00000127371
ENSDART00000100276 |
tmco1
|
transmembrane and coiled-coil domains 1 |
| chr4_-_77130289 | 0.73 |
ENSDART00000174380
|
CU467646.7
|
|
| chr12_+_21697540 | 0.73 |
ENSDART00000153412
|
pkd1b
|
polycystic kidney disease 1b |
| chr3_+_31662126 | 0.72 |
ENSDART00000113441
|
mylk5
|
myosin, light chain kinase 5 |
| chr11_-_6974022 | 0.72 |
ENSDART00000172851
|
COMP
|
si:ch211-43f4.1 |
| chr10_+_8875195 | 0.69 |
ENSDART00000141045
|
itga2.3
|
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor), tandem duplicate 3 |
| chr5_-_41860556 | 0.69 |
ENSDART00000140154
|
si:dkey-65b12.12
|
si:dkey-65b12.12 |
| chr15_+_5973909 | 0.68 |
ENSDART00000126886
ENSDART00000189618 |
igsf5b
|
immunoglobulin superfamily, member 5b |
| chr2_-_7246848 | 0.68 |
ENSDART00000146434
|
zgc:153115
|
zgc:153115 |
| chr10_-_29892486 | 0.67 |
ENSDART00000099983
|
bsx
|
brain-specific homeobox |
| chr11_-_40681011 | 0.67 |
ENSDART00000166372
ENSDART00000110622 ENSDART00000159713 |
si:ch211-222l21.1
|
si:ch211-222l21.1 |
| chr1_-_58868306 | 0.66 |
ENSDART00000166615
|
dnm2b
|
dynamin 2b |
| chr21_-_5879897 | 0.66 |
ENSDART00000184034
|
rpl35
|
ribosomal protein L35 |
| chr4_-_77135076 | 0.65 |
ENSDART00000174184
|
zgc:173770
|
zgc:173770 |
| chr5_+_69650148 | 0.65 |
ENSDART00000097244
|
gtf2h3
|
general transcription factor IIH, polypeptide 3 |
| chr2_+_44977889 | 0.65 |
ENSDART00000144024
|
alg3
|
asparagine-linked glycosylation 3 (alpha-1,3-mannosyltransferase) |
| chr3_+_30501135 | 0.64 |
ENSDART00000165869
|
si:dkey-13n23.3
|
si:dkey-13n23.3 |
| chr15_-_23206562 | 0.64 |
ENSDART00000110360
|
ccdc153
|
coiled-coil domain containing 153 |
| chr7_+_38267136 | 0.64 |
ENSDART00000173613
|
gpatch1
|
G patch domain containing 1 |
| chr22_-_5252005 | 0.64 |
ENSDART00000132942
ENSDART00000081801 |
ncln
|
nicalin |
| chr2_-_41562868 | 0.64 |
ENSDART00000084597
|
d2hgdh
|
D-2-hydroxyglutarate dehydrogenase |
| chr23_-_24488696 | 0.63 |
ENSDART00000155593
|
tmem82
|
transmembrane protein 82 |
| chr22_+_465269 | 0.62 |
ENSDART00000145767
|
celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr20_+_49081967 | 0.61 |
ENSDART00000112689
|
crnkl1
|
crooked neck pre-mRNA splicing factor 1 |
| chr2_-_36658701 | 0.61 |
ENSDART00000184995
|
CT027583.2
|
|
| chr14_+_30413758 | 0.60 |
ENSDART00000092953
|
cnot7
|
CCR4-NOT transcription complex, subunit 7 |
| chr25_+_3525494 | 0.60 |
ENSDART00000169775
|
RAB19 (1 of many)
|
RAB19, member RAS oncogene family |
| chr4_-_77135340 | 0.60 |
ENSDART00000180581
ENSDART00000179901 |
CU467646.7
|
|
| chr20_-_36887516 | 0.60 |
ENSDART00000076313
|
txlnba
|
taxilin beta a |
| chr4_-_77116266 | 0.60 |
ENSDART00000174249
|
CU467646.2
|
|
| chr2_-_56649883 | 0.58 |
ENSDART00000191786
|
gpx4b
|
glutathione peroxidase 4b |
| chr10_+_24692076 | 0.58 |
ENSDART00000181600
|
tpte
|
transmembrane phosphatase with tensin homology |
| chr5_-_3839285 | 0.57 |
ENSDART00000122292
|
mlxipl
|
MLX interacting protein like |
| chr17_-_18888959 | 0.57 |
ENSDART00000080029
|
ak7b
|
adenylate kinase 7b |
| chr11_-_7410537 | 0.57 |
ENSDART00000009859
|
adgrl4
|
adhesion G protein-coupled receptor L4 |
| chr13_+_31497236 | 0.56 |
ENSDART00000146752
|
lrrc9
|
leucine rich repeat containing 9 |
| chr1_-_51720633 | 0.56 |
ENSDART00000045894
|
rnaseh2a
|
ribonuclease H2, subunit A |
| chr23_-_27607039 | 0.56 |
ENSDART00000183639
|
phf8
|
PHD finger protein 8 |
| chr6_-_40044111 | 0.55 |
ENSDART00000154347
|
si:dkey-197j19.5
|
si:dkey-197j19.5 |
| chr10_+_20364009 | 0.54 |
ENSDART00000186139
ENSDART00000080395 |
golga7
|
golgin A7 |
| chr6_+_30533504 | 0.54 |
ENSDART00000155842
|
wwc3
|
WWC family member 3 |
| chr17_-_25303486 | 0.54 |
ENSDART00000162235
|
ppie
|
peptidylprolyl isomerase E (cyclophilin E) |
| chr20_+_19066858 | 0.53 |
ENSDART00000192086
|
sox7
|
SRY (sex determining region Y)-box 7 |
| chr12_+_45238292 | 0.52 |
ENSDART00000057983
|
mrpl38
|
mitochondrial ribosomal protein L38 |
| chr24_+_13869092 | 0.52 |
ENSDART00000176492
|
eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr1_-_52498146 | 0.52 |
ENSDART00000122217
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr15_+_40188076 | 0.52 |
ENSDART00000063779
|
efhd1
|
EF-hand domain family, member D1 |
| chr22_+_15720381 | 0.52 |
ENSDART00000128149
|
fam32a
|
family with sequence similarity 32, member A |
| chr19_+_20787179 | 0.51 |
ENSDART00000193204
|
adnp2b
|
ADNP homeobox 2b |
| chr21_+_261490 | 0.51 |
ENSDART00000177919
|
jak2a
|
Janus kinase 2a |
| chr20_-_36887176 | 0.50 |
ENSDART00000143515
ENSDART00000146994 |
txlnba
|
taxilin beta a |
| chr23_+_39346930 | 0.50 |
ENSDART00000102843
|
src
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr23_-_9855627 | 0.50 |
ENSDART00000180159
|
prkcbp1l
|
protein kinase C binding protein 1, like |
| chr7_-_32021853 | 0.49 |
ENSDART00000134521
|
kif18a
|
kinesin family member 18A |
| chr13_+_7241170 | 0.49 |
ENSDART00000109434
|
aifm2
|
apoptosis-inducing factor, mitochondrion-associated, 2 |
| chr6_-_35046735 | 0.49 |
ENSDART00000143649
|
uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr19_-_47997424 | 0.49 |
ENSDART00000081675
|
ctnnb2
|
catenin, beta 2 |
| chr21_-_15200556 | 0.49 |
ENSDART00000141809
|
sfswap
|
splicing factor SWAP |
| chr6_+_30703828 | 0.48 |
ENSDART00000104628
|
insl5a
|
insulin-like 5a |
| chr13_+_43247936 | 0.48 |
ENSDART00000126850
ENSDART00000165331 |
smoc2
|
SPARC related modular calcium binding 2 |
| chr10_+_36441124 | 0.47 |
ENSDART00000185626
|
uspl1
|
ubiquitin specific peptidase like 1 |
| chr3_-_34561624 | 0.47 |
ENSDART00000129313
|
sept9a
|
septin 9a |
| chr3_+_30500968 | 0.47 |
ENSDART00000103447
|
si:dkey-13n23.3
|
si:dkey-13n23.3 |
| chr19_+_2631565 | 0.47 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr19_+_2877079 | 0.46 |
ENSDART00000160847
|
hgh1
|
HGH1 homolog (S. cerevisiae) |
| chr4_-_4535189 | 0.46 |
ENSDART00000057519
|
zgc:194209
|
zgc:194209 |
| chr24_+_81527 | 0.46 |
ENSDART00000192139
|
reck
|
reversion-inducing-cysteine-rich protein with kazal motifs |
| chr9_-_33477588 | 0.45 |
ENSDART00000144150
|
caska
|
calcium/calmodulin-dependent serine protein kinase a |
| chr14_-_46675850 | 0.45 |
ENSDART00000113285
|
tapt1a
|
transmembrane anterior posterior transformation 1a |
| chr3_-_3448095 | 0.45 |
ENSDART00000078886
|
A2ML1 (1 of many)
|
si:dkey-46g23.5 |
| chr24_-_26820698 | 0.44 |
ENSDART00000147788
|
fndc3bb
|
fibronectin type III domain containing 3Bb |
| chr19_+_11214007 | 0.44 |
ENSDART00000127362
|
si:ch73-109i22.2
|
si:ch73-109i22.2 |
| chr25_-_10622449 | 0.44 |
ENSDART00000155375
|
ppp6r3
|
protein phosphatase 6, regulatory subunit 3 |
| chr14_-_48103207 | 0.44 |
ENSDART00000056712
|
etfdh
|
electron-transferring-flavoprotein dehydrogenase |
| chr20_+_19066596 | 0.44 |
ENSDART00000130271
|
sox7
|
SRY (sex determining region Y)-box 7 |
| chr10_-_25591194 | 0.44 |
ENSDART00000131640
|
tiam1a
|
T cell lymphoma invasion and metastasis 1a |
| chr9_-_43142636 | 0.43 |
ENSDART00000134349
ENSDART00000181835 |
ccdc141
|
coiled-coil domain containing 141 |
| chr15_-_25365570 | 0.43 |
ENSDART00000152754
|
cluha
|
clustered mitochondria (cluA/CLU1) homolog a |
| chr23_+_39346774 | 0.43 |
ENSDART00000190985
|
src
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr3_-_26921475 | 0.43 |
ENSDART00000130281
|
ciita
|
class II, major histocompatibility complex, transactivator |
| chr2_-_3045861 | 0.43 |
ENSDART00000105818
ENSDART00000187575 |
guk1a
|
guanylate kinase 1a |
| chr9_+_23770666 | 0.42 |
ENSDART00000182493
|
si:ch211-219a4.3
|
si:ch211-219a4.3 |
| chr7_+_22586800 | 0.42 |
ENSDART00000035325
|
chrnb1l
|
cholinergic receptor, nicotinic, beta 1 (muscle) like |
| chr25_+_21829777 | 0.42 |
ENSDART00000027393
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr7_-_19146925 | 0.42 |
ENSDART00000142924
ENSDART00000009695 |
kirrel1a
|
kirre like nephrin family adhesion molecule 1a |
| chr9_-_29985390 | 0.42 |
ENSDART00000134157
|
il1rapl1a
|
interleukin 1 receptor accessory protein-like 1a |
| chr16_+_17252487 | 0.42 |
ENSDART00000063572
|
gtf3c6
|
general transcription factor IIIC, polypeptide 6, alpha |
| chr24_+_22022109 | 0.41 |
ENSDART00000133686
|
ropn1l
|
rhophilin associated tail protein 1-like |
| chr5_+_29831235 | 0.41 |
ENSDART00000109660
|
f11r.1
|
F11 receptor, tandem duplicate 1 |
| chr5_+_52167986 | 0.41 |
ENSDART00000162256
ENSDART00000073626 |
slc14a2
|
solute carrier family 14 (urea transporter), member 2 |
| chr14_+_20941534 | 0.40 |
ENSDART00000185616
|
zgc:66433
|
zgc:66433 |
| chr7_-_26924903 | 0.39 |
ENSDART00000124363
|
alx4a
|
ALX homeobox 4a |
| chr9_+_41914378 | 0.39 |
ENSDART00000130434
ENSDART00000007058 |
col18a1a
|
collagen type XVIII alpha 1 chain a |
| chr15_+_11840311 | 0.38 |
ENSDART00000167671
|
prkd2
|
protein kinase D2 |
| chr23_+_24272421 | 0.38 |
ENSDART00000029974
|
clcnk
|
chloride channel K |
| chr6_-_54111928 | 0.38 |
ENSDART00000083880
|
hyal2a
|
hyaluronoglucosaminidase 2a |
| chr1_-_17650223 | 0.38 |
ENSDART00000043484
|
si:dkey-256e7.5
|
si:dkey-256e7.5 |
| chr22_+_29113796 | 0.38 |
ENSDART00000150264
|
pla2g6
|
phospholipase A2, group VI (cytosolic, calcium-independent) |
| chr8_-_52909850 | 0.36 |
ENSDART00000161943
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr16_-_19568795 | 0.36 |
ENSDART00000185141
|
abcb5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr22_-_15720347 | 0.36 |
ENSDART00000105692
|
mrpl54
|
mitochondrial ribosomal protein L54 |
| chr12_+_23866368 | 0.36 |
ENSDART00000188652
ENSDART00000192478 |
svila
|
supervillin a |
| chr16_+_25342152 | 0.36 |
ENSDART00000156351
|
zfat
|
zinc finger and AT hook domain containing |
| chr12_+_47439 | 0.36 |
ENSDART00000114732
|
mrpl58
|
mitochondrial ribosomal protein L58 |
| chr20_-_25877793 | 0.36 |
ENSDART00000177333
ENSDART00000184619 |
exoc1
|
exocyst complex component 1 |
| chr1_-_52497834 | 0.36 |
ENSDART00000136469
ENSDART00000004233 |
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr12_+_46791907 | 0.35 |
ENSDART00000110304
|
vcla
|
vinculin a |
| chr9_+_23772516 | 0.35 |
ENSDART00000183126
|
si:ch211-219a4.3
|
si:ch211-219a4.3 |
| chr4_-_47304459 | 0.35 |
ENSDART00000169561
|
si:ch211-261d9.1
|
si:ch211-261d9.1 |
| chr4_-_77120928 | 0.35 |
ENSDART00000174154
|
CU467646.1
|
|
| chr7_-_71331499 | 0.35 |
ENSDART00000081245
|
dhx15
|
DEAH (Asp-Glu-Ala-His) box helicase 15 |
| chr2_-_31700396 | 0.35 |
ENSDART00000142366
|
lrrcc1
|
leucine rich repeat and coiled-coil centrosomal protein 1 |
| chr3_+_3454610 | 0.34 |
ENSDART00000024900
|
zgc:165453
|
zgc:165453 |
| chr5_+_24156170 | 0.34 |
ENSDART00000136570
|
slc25a15b
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15b |
| chr23_-_20154422 | 0.33 |
ENSDART00000132025
|
usp19
|
ubiquitin specific peptidase 19 |
| chr1_+_13087797 | 0.33 |
ENSDART00000170314
|
si:dkey-5n7.2
|
si:dkey-5n7.2 |
| chr9_+_9502610 | 0.32 |
ENSDART00000061525
ENSDART00000125174 |
nr1i2
|
nuclear receptor subfamily 1, group I, member 2 |
| chr12_-_34827477 | 0.32 |
ENSDART00000153026
|
NDUFAF8
|
si:dkey-21c1.6 |
| chr3_+_35611625 | 0.32 |
ENSDART00000190995
|
traf7
|
TNF receptor-associated factor 7 |
| chr20_+_18260358 | 0.32 |
ENSDART00000187734
ENSDART00000191947 ENSDART00000057039 |
taf4b
|
TAF4B RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr15_-_6615555 | 0.32 |
ENSDART00000152725
|
atm
|
ATM serine/threonine kinase |
| chr1_-_28861226 | 0.31 |
ENSDART00000075502
|
timmdc1
|
translocase of inner mitochondrial membrane domain containing 1 |
| chr17_-_46850299 | 0.30 |
ENSDART00000154430
|
pimr23
|
Pim proto-oncogene, serine/threonine kinase, related 23 |
| chr18_-_48492951 | 0.30 |
ENSDART00000146346
|
kcnj1a.6
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 6 |
| chr9_+_35077546 | 0.30 |
ENSDART00000142688
|
si:dkey-192g7.3
|
si:dkey-192g7.3 |
| chr2_+_30249977 | 0.30 |
ENSDART00000109160
ENSDART00000135171 |
tmem70
|
transmembrane protein 70 |
| chr3_+_57825938 | 0.30 |
ENSDART00000128815
|
cenpx
|
centromere protein X |
| chr15_-_16946124 | 0.30 |
ENSDART00000154923
|
hip1
|
huntingtin interacting protein 1 |
| chr12_-_29305533 | 0.29 |
ENSDART00000189410
|
sh2d4bb
|
SH2 domain containing 4Bb |
| chr24_-_1021318 | 0.29 |
ENSDART00000181403
|
ralaa
|
v-ral simian leukemia viral oncogene homolog Aa (ras related) |
| chr18_-_43866526 | 0.28 |
ENSDART00000111309
|
treh
|
trehalase (brush-border membrane glycoprotein) |
| chr19_-_24136233 | 0.28 |
ENSDART00000143365
|
thap7
|
THAP domain containing 7 |
| chr20_+_37825804 | 0.28 |
ENSDART00000152865
|
tatdn3
|
TatD DNase domain containing 3 |
| chr8_-_7440364 | 0.28 |
ENSDART00000180659
|
hdac6
|
histone deacetylase 6 |
| chr14_+_30413312 | 0.28 |
ENSDART00000186864
|
cnot7
|
CCR4-NOT transcription complex, subunit 7 |
| chr15_-_16884912 | 0.28 |
ENSDART00000062135
|
zgc:103681
|
zgc:103681 |
| chr14_-_17121676 | 0.27 |
ENSDART00000170154
ENSDART00000060479 |
smtnl1
|
smoothelin-like 1 |
| chr12_-_4185554 | 0.27 |
ENSDART00000152313
|
si:dkey-32n7.7
|
si:dkey-32n7.7 |
| chr3_-_7129057 | 0.27 |
ENSDART00000125947
|
BX005085.1
|
|
| chr5_+_37406358 | 0.26 |
ENSDART00000162811
|
klhl13
|
kelch-like family member 13 |
| chr10_+_8629275 | 0.26 |
ENSDART00000129643
|
aplnrb
|
apelin receptor b |
| chr17_+_450956 | 0.26 |
ENSDART00000183022
ENSDART00000171386 |
zgc:194887
|
zgc:194887 |
| chr3_-_34034498 | 0.26 |
ENSDART00000151145
|
ighv11-2
|
immunoglobulin heavy variable 11-2 |
| chr16_-_19349318 | 0.25 |
ENSDART00000020821
|
dnah11
|
dynein, axonemal, heavy chain 11 |
| chr13_-_44842884 | 0.25 |
ENSDART00000184195
|
si:dkeyp-2e4.3
|
si:dkeyp-2e4.3 |
| chr3_-_32590164 | 0.25 |
ENSDART00000151151
|
tspan4b
|
tetraspanin 4b |
| chr1_-_14694809 | 0.25 |
ENSDART00000193331
|
CU138548.5
|
|
| chr16_-_30927454 | 0.25 |
ENSDART00000184015
|
slc45a4
|
solute carrier family 45, member 4 |
| chr16_+_23913943 | 0.24 |
ENSDART00000175404
ENSDART00000129525 |
apoa4b.1
|
apolipoprotein A-IV b, tandem duplicate 1 |
| chr6_+_52924103 | 0.24 |
ENSDART00000065693
|
or137-2
|
odorant receptor, family H, subfamily 137, member 2 |
| chr7_+_29012033 | 0.24 |
ENSDART00000173909
ENSDART00000145762 |
dnaaf1
|
dynein, axonemal, assembly factor 1 |
| chr3_+_62317990 | 0.23 |
ENSDART00000058719
|
znf1124
|
zinc finger protein 1124 |
| chr10_+_26645953 | 0.23 |
ENSDART00000131482
|
adgrg4b
|
adhesion G protein-coupled receptor G4b |
| chr19_+_1465004 | 0.23 |
ENSDART00000159157
|
CABZ01073736.1
|
|
| chr16_+_20871021 | 0.23 |
ENSDART00000006429
|
hibadhb
|
3-hydroxyisobutyrate dehydrogenase b |
| chr24_-_2843107 | 0.22 |
ENSDART00000165290
|
cyb5a
|
cytochrome b5 type A (microsomal) |
| chr3_+_12816362 | 0.22 |
ENSDART00000163743
ENSDART00000170788 |
cyp2k6
|
cytochrome P450, family 2, subfamily K, polypeptide 6 |
| chr16_-_30655980 | 0.22 |
ENSDART00000146508
|
ldlrad4b
|
low density lipoprotein receptor class A domain containing 4b |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr4a3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.4 | 2.2 | GO:0009219 | pyrimidine deoxyribonucleotide metabolic process(GO:0009219) |
| 0.4 | 1.7 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.4 | 2.1 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.3 | 0.9 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.3 | 0.8 | GO:0036363 | transforming growth factor beta activation(GO:0036363) |
| 0.2 | 1.9 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.2 | 0.6 | GO:0061187 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.2 | 0.7 | GO:0032965 | regulation of collagen metabolic process(GO:0010712) regulation of collagen biosynthetic process(GO:0032965) regulation of multicellular organismal metabolic process(GO:0044246) |
| 0.2 | 0.5 | GO:1905048 | regulation of metallopeptidase activity(GO:1905048) |
| 0.2 | 1.1 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.1 | 0.4 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.1 | 1.5 | GO:0035306 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.1 | 0.6 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.1 | 0.4 | GO:0050787 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 0.1 | 0.4 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.1 | 0.3 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.1 | 0.7 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.1 | 0.3 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.1 | 0.7 | GO:0021982 | pineal gland development(GO:0021982) |
| 0.1 | 0.5 | GO:0030576 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) protein localization to nucleoplasm(GO:1990173) |
| 0.1 | 0.3 | GO:0005991 | trehalose metabolic process(GO:0005991) |
| 0.1 | 0.6 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 0.5 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.9 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.1 | 2.4 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.1 | 0.2 | GO:0046222 | mycotoxin metabolic process(GO:0043385) aflatoxin metabolic process(GO:0046222) organic heteropentacyclic compound metabolic process(GO:1901376) |
| 0.1 | 0.8 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.1 | 0.3 | GO:1903589 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903589) |
| 0.1 | 0.4 | GO:0089700 | endocardial cushion formation(GO:0003272) protein kinase D signaling(GO:0089700) |
| 0.1 | 0.7 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.1 | 0.4 | GO:0086013 | membrane repolarization during cardiac muscle cell action potential(GO:0086013) |
| 0.1 | 0.5 | GO:1901099 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.1 | 0.3 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.1 | 0.4 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.6 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.0 | 0.6 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.2 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.7 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.4 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.8 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.4 | GO:0006603 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 0.1 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.0 | 0.5 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.2 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.4 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.0 | 0.4 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.3 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.0 | 0.2 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.4 | GO:0045920 | negative regulation of exocytosis(GO:0045920) |
| 0.0 | 3.4 | GO:0006979 | response to oxidative stress(GO:0006979) |
| 0.0 | 0.1 | GO:0006549 | isoleucine metabolic process(GO:0006549) isoleucine biosynthetic process(GO:0009097) |
| 0.0 | 1.6 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.5 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.4 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 0.7 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.2 | GO:0003314 | heart rudiment morphogenesis(GO:0003314) motile cilium assembly(GO:0044458) |
| 0.0 | 0.9 | GO:0010906 | regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 0.4 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.8 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.2 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 1.0 | GO:0060840 | artery development(GO:0060840) |
| 0.0 | 0.5 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 0.5 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:1903817 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.3 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.5 | GO:0008637 | apoptotic mitochondrial changes(GO:0008637) |
| 0.0 | 0.5 | GO:1904894 | positive regulation of JAK-STAT cascade(GO:0046427) positive regulation of STAT cascade(GO:1904894) |
| 0.0 | 0.5 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.4 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.3 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 1.2 | GO:1902593 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 0.3 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.0 | 0.2 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.7 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.4 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.2 | GO:0003209 | cardiac atrium morphogenesis(GO:0003209) |
| 0.0 | 0.1 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0034244 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.6 | GO:0051896 | regulation of protein kinase B signaling(GO:0051896) |
| 0.0 | 0.6 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.2 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.2 | GO:2000404 | regulation of T cell migration(GO:2000404) |
| 0.0 | 0.4 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.0 | 0.3 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 | 0.1 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.0 | 0.5 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.1 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 0.0 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 0.6 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 0.5 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.1 | 0.5 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 0.6 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 1.2 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.1 | 0.3 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.1 | 0.4 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 0.6 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.1 | 0.8 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.9 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.5 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.8 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.1 | GO:0005948 | acetolactate synthase complex(GO:0005948) |
| 0.0 | 0.5 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 1.2 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.3 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.2 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 0.8 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.8 | GO:0015934 | large ribosomal subunit(GO:0015934) |
| 0.0 | 0.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.5 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.3 | 0.9 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.3 | 1.2 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.2 | 2.1 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.2 | 0.9 | GO:0016519 | gastric inhibitory peptide receptor activity(GO:0016519) |
| 0.2 | 1.0 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.2 | 0.6 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.2 | 0.8 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.2 | 1.1 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.1 | 0.4 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.1 | 0.6 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 1.4 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.1 | 0.6 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.1 | 1.8 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.5 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.1 | 0.6 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 0.4 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.1 | 0.5 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 0.3 | GO:0004555 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 0.1 | 0.4 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 0.9 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 0.6 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 1.0 | GO:0002020 | protease binding(GO:0002020) |
| 0.1 | 0.3 | GO:0047611 | tubulin deacetylase activity(GO:0042903) acetylspermidine deacetylase activity(GO:0047611) |
| 0.1 | 1.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.2 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 0.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.2 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.1 | 0.2 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.1 | 0.9 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.7 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.5 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.3 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.0 | 0.4 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.2 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 0.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.5 | GO:0016840 | carbon-nitrogen lyase activity(GO:0016840) |
| 0.0 | 0.3 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.4 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.5 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0003984 | acetolactate synthase activity(GO:0003984) |
| 0.0 | 0.4 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 1.2 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 1.2 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.0 | 0.2 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.0 | 0.3 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.4 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.2 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.2 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.3 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 0.7 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.9 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 0.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.9 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.1 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 2.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.9 | GO:0070851 | growth factor receptor binding(GO:0070851) |
| 0.0 | 0.2 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 1.1 | GO:0019905 | syntaxin binding(GO:0019905) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 1.1 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 1.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.1 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 2.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.3 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 0.9 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 1.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.2 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 0.8 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.3 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.6 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.4 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.8 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.4 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 1.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.4 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |