Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
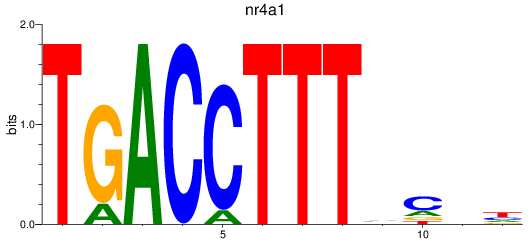
Results for nr4a1
Z-value: 0.89
Transcription factors associated with nr4a1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr4a1
|
ENSDARG00000000796 | nuclear receptor subfamily 4, group A, member 1 |
|
nr4a1
|
ENSDARG00000109473 | nuclear receptor subfamily 4, group A, member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr4a1 | dr11_v1_chr23_-_32156278_32156278 | 0.57 | 1.0e-02 | Click! |
Activity profile of nr4a1 motif
Sorted Z-values of nr4a1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_11385155 | 5.45 |
ENSDART00000064214
|
plac8.1
|
placenta-specific 8, tandem duplicate 1 |
| chr5_+_1278092 | 5.20 |
ENSDART00000147972
ENSDART00000159783 |
dnm1a
|
dynamin 1a |
| chr19_-_703898 | 3.44 |
ENSDART00000181096
ENSDART00000121462 |
slc6a19a.2
|
solute carrier family 6 (neutral amino acid transporter), member 19a, tandem duplicate 2 |
| chr23_-_45504991 | 2.98 |
ENSDART00000148761
|
col24a1
|
collagen type XXIV alpha 1 |
| chr5_+_45677781 | 2.97 |
ENSDART00000163120
ENSDART00000126537 |
gc
|
group-specific component (vitamin D binding protein) |
| chr1_-_50838160 | 2.79 |
ENSDART00000163939
ENSDART00000165111 |
zgc:154142
|
zgc:154142 |
| chr15_-_12270857 | 2.76 |
ENSDART00000170093
|
si:dkey-36i7.3
|
si:dkey-36i7.3 |
| chr25_+_29160102 | 2.68 |
ENSDART00000162854
|
pkmb
|
pyruvate kinase M1/2b |
| chr8_-_14052349 | 2.10 |
ENSDART00000135811
|
atp2b3a
|
ATPase plasma membrane Ca2+ transporting 3a |
| chr20_-_14206698 | 1.98 |
ENSDART00000082441
|
dlgap2b
|
discs, large (Drosophila) homolog-associated protein 2b |
| chr6_-_31348999 | 1.97 |
ENSDART00000153734
|
dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr10_-_17745345 | 1.95 |
ENSDART00000132690
ENSDART00000135376 |
si:dkey-200l5.4
|
si:dkey-200l5.4 |
| chr3_+_30257582 | 1.90 |
ENSDART00000159497
ENSDART00000103457 ENSDART00000121883 |
mybpc2a
|
myosin binding protein C, fast type a |
| chr5_-_24238733 | 1.82 |
ENSDART00000138170
|
plscr3a
|
phospholipid scramblase 3a |
| chr6_-_10320676 | 1.77 |
ENSDART00000151247
|
scn1lab
|
sodium channel, voltage-gated, type I like, alpha b |
| chr1_+_44826593 | 1.74 |
ENSDART00000162200
|
STX3
|
zgc:165520 |
| chr6_-_53204808 | 1.72 |
ENSDART00000160955
|
slc38a3b
|
solute carrier family 38, member 3b |
| chr1_+_19332837 | 1.68 |
ENSDART00000078594
|
tyrp1b
|
tyrosinase-related protein 1b |
| chr21_+_10076203 | 1.65 |
ENSDART00000190383
|
CABZ01043506.1
|
|
| chr7_-_39552314 | 1.63 |
ENSDART00000134174
|
slc22a18
|
solute carrier family 22, member 18 |
| chr22_-_16997475 | 1.62 |
ENSDART00000090237
|
nfia
|
nuclear factor I/A |
| chr6_-_49873020 | 1.59 |
ENSDART00000148511
|
gnas
|
GNAS complex locus |
| chr18_-_15771551 | 1.59 |
ENSDART00000130931
ENSDART00000154079 |
si:ch211-219a15.3
|
si:ch211-219a15.3 |
| chr7_+_34620418 | 1.58 |
ENSDART00000081338
|
slc9a5
|
solute carrier family 9, subfamily A (NHE5, cation proton antiporter 5), member 5 |
| chr7_-_42206720 | 1.52 |
ENSDART00000110907
|
itfg1
|
integrin alpha FG-GAP repeat containing 1 |
| chr7_-_39551913 | 1.44 |
ENSDART00000173498
ENSDART00000143040 |
slc22a18
|
solute carrier family 22, member 18 |
| chr14_+_14225048 | 1.44 |
ENSDART00000168749
|
nlgn3a
|
neuroligin 3a |
| chr2_-_11912347 | 1.43 |
ENSDART00000023851
|
abhd3
|
abhydrolase domain containing 3 |
| chr13_-_21739142 | 1.40 |
ENSDART00000078460
|
si:dkey-191g9.5
|
si:dkey-191g9.5 |
| chr5_+_19448078 | 1.39 |
ENSDART00000088968
|
ube3b
|
ubiquitin protein ligase E3B |
| chr14_+_14224730 | 1.35 |
ENSDART00000180112
ENSDART00000184891 ENSDART00000174760 |
nlgn3a
|
neuroligin 3a |
| chr5_-_36549024 | 1.35 |
ENSDART00000097671
|
zgc:158432
|
zgc:158432 |
| chr6_+_3282809 | 1.29 |
ENSDART00000187444
ENSDART00000187407 ENSDART00000191883 |
kdm4aa
|
lysine (K)-specific demethylase 4A, genome duplicate a |
| chr10_+_41668483 | 1.21 |
ENSDART00000127073
|
lrrc75bb
|
leucine rich repeat containing 75Bb |
| chr11_-_17713987 | 1.21 |
ENSDART00000090401
|
fam19a4b
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A4b |
| chr11_+_1409622 | 1.20 |
ENSDART00000157042
|
ppp1r3db
|
protein phosphatase 1, regulatory subunit 3Db |
| chr23_+_21544227 | 1.17 |
ENSDART00000140253
|
arhgef10lb
|
Rho guanine nucleotide exchange factor (GEF) 10-like b |
| chr10_-_25699454 | 1.11 |
ENSDART00000064376
|
sod1
|
superoxide dismutase 1, soluble |
| chr8_+_26125218 | 1.10 |
ENSDART00000145095
|
celsr3
|
cadherin, EGF LAG seven-pass G-type receptor 3 |
| chr21_-_30648106 | 1.10 |
ENSDART00000160800
ENSDART00000177022 |
phka1b
|
phosphorylase kinase, alpha 1b (muscle) |
| chr21_-_43527198 | 1.08 |
ENSDART00000126092
|
irs4a
|
insulin receptor substrate 4a |
| chr3_-_37351225 | 1.07 |
ENSDART00000174685
|
si:ch211-278a6.1
|
si:ch211-278a6.1 |
| chr10_+_7563755 | 1.05 |
ENSDART00000165877
|
purg
|
purine-rich element binding protein G |
| chr12_-_31457301 | 1.04 |
ENSDART00000043887
ENSDART00000148603 |
acsl5
|
acyl-CoA synthetase long chain family member 5 |
| chr8_-_45835056 | 1.04 |
ENSDART00000022242
|
ogdha
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase a (lipoamide) |
| chr5_+_62611400 | 1.04 |
ENSDART00000132054
|
abr
|
active BCR-related |
| chr12_-_6880694 | 1.04 |
ENSDART00000171846
|
pcdh15b
|
protocadherin-related 15b |
| chr8_+_10404310 | 1.03 |
ENSDART00000185573
|
tbc1d22b
|
TBC1 domain family, member 22B |
| chr7_+_44484853 | 1.01 |
ENSDART00000189079
ENSDART00000121826 |
bean1
|
brain expressed, associated with NEDD4, 1 |
| chr7_+_42206847 | 1.01 |
ENSDART00000149250
|
phkb
|
phosphorylase kinase, beta |
| chr7_-_24005268 | 1.00 |
ENSDART00000173608
|
si:dkey-183c6.9
|
si:dkey-183c6.9 |
| chr19_+_3056450 | 0.96 |
ENSDART00000141324
ENSDART00000082353 |
hsf1
|
heat shock transcription factor 1 |
| chr17_-_7371564 | 0.95 |
ENSDART00000060336
|
rab32b
|
RAB32b, member RAS oncogene family |
| chr18_-_2433011 | 0.93 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr16_+_19677484 | 0.93 |
ENSDART00000150588
|
tmem196b
|
transmembrane protein 196b |
| chr1_-_23110740 | 0.92 |
ENSDART00000171848
ENSDART00000086797 ENSDART00000189344 ENSDART00000190858 |
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr7_+_17229282 | 0.91 |
ENSDART00000097982
|
slc6a5
|
solute carrier family 6 (neurotransmitter transporter), member 5 |
| chr15_+_28268135 | 0.90 |
ENSDART00000152536
ENSDART00000188550 |
myo1cb
|
myosin Ic, paralog b |
| chr20_+_46897504 | 0.89 |
ENSDART00000158124
|
si:ch73-21k16.1
|
si:ch73-21k16.1 |
| chr3_-_5829501 | 0.88 |
ENSDART00000091017
|
pkn1b
|
protein kinase N1b |
| chr18_+_27515640 | 0.88 |
ENSDART00000181593
|
tp53i11b
|
tumor protein p53 inducible protein 11b |
| chr6_-_35738836 | 0.86 |
ENSDART00000111642
|
brinp3a.1
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3a, tandem duplicate 1 |
| chr25_-_31898552 | 0.86 |
ENSDART00000156128
|
si:ch73-330k17.3
|
si:ch73-330k17.3 |
| chr7_+_56253914 | 0.84 |
ENSDART00000073594
|
ankrd11
|
ankyrin repeat domain 11 |
| chr22_+_21317597 | 0.83 |
ENSDART00000132605
|
shc2
|
SHC (Src homology 2 domain containing) transforming protein 2 |
| chr11_+_11201096 | 0.82 |
ENSDART00000171916
ENSDART00000171521 ENSDART00000087105 ENSDART00000159603 |
myom2a
|
myomesin 2a |
| chr12_+_24562667 | 0.81 |
ENSDART00000056256
|
nrxn1a
|
neurexin 1a |
| chr14_+_36628131 | 0.81 |
ENSDART00000188625
ENSDART00000125345 |
TENM3
|
si:dkey-237h12.3 |
| chr20_+_40150612 | 0.81 |
ENSDART00000143680
ENSDART00000109681 ENSDART00000101041 ENSDART00000121818 |
trdn
|
triadin |
| chr21_+_23953181 | 0.79 |
ENSDART00000145541
ENSDART00000065599 ENSDART00000112869 |
cadm1a
|
cell adhesion molecule 1a |
| chr23_+_3538463 | 0.77 |
ENSDART00000172758
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr16_-_22294265 | 0.77 |
ENSDART00000124718
|
aqp10a
|
aquaporin 10a |
| chr9_-_30346279 | 0.76 |
ENSDART00000089488
|
sytl5
|
synaptotagmin-like 5 |
| chr7_-_24046999 | 0.76 |
ENSDART00000144616
ENSDART00000124653 ENSDART00000127813 |
dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr18_+_50461981 | 0.75 |
ENSDART00000158761
|
CU896640.1
|
|
| chr7_+_34297271 | 0.74 |
ENSDART00000180342
|
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr10_+_7564106 | 0.71 |
ENSDART00000159042
|
purg
|
purine-rich element binding protein G |
| chr18_+_31410652 | 0.70 |
ENSDART00000098504
|
def8
|
differentially expressed in FDCP 8 homolog (mouse) |
| chr2_+_10127762 | 0.70 |
ENSDART00000100726
|
insl5b
|
insulin-like 5b |
| chr11_-_44999858 | 0.70 |
ENSDART00000167759
ENSDART00000126845 |
ldb1b
|
LIM-domain binding 1b |
| chr5_+_65493699 | 0.68 |
ENSDART00000161567
|
si:dkey-21e5.1
|
si:dkey-21e5.1 |
| chr16_-_16619854 | 0.65 |
ENSDART00000150512
ENSDART00000191306 ENSDART00000181773 ENSDART00000183231 |
cyp21a2
|
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chr3_-_30388525 | 0.64 |
ENSDART00000183790
|
lrrc4ba
|
leucine rich repeat containing 4Ba |
| chr18_-_42333428 | 0.64 |
ENSDART00000034225
|
cntn5
|
contactin 5 |
| chr17_+_5800286 | 0.63 |
ENSDART00000175504
|
rp1l1a
|
retinitis pigmentosa 1-like 1a |
| chr12_+_46708920 | 0.61 |
ENSDART00000153089
|
exoc7
|
exocyst complex component 7 |
| chr12_+_21525496 | 0.61 |
ENSDART00000152974
|
ca10a
|
carbonic anhydrase Xa |
| chr25_+_6266009 | 0.60 |
ENSDART00000148995
|
slc25a44a
|
solute carrier family 25, member 44 a |
| chr4_-_7212875 | 0.59 |
ENSDART00000161297
|
lrrn3b
|
leucine rich repeat neuronal 3b |
| chr8_+_10823069 | 0.59 |
ENSDART00000081341
|
mapk13
|
mitogen-activated protein kinase 13 |
| chr12_+_9817440 | 0.58 |
ENSDART00000137081
ENSDART00000123712 |
rundc3ab
|
RUN domain containing 3Ab |
| chr6_-_39764995 | 0.57 |
ENSDART00000085277
|
pfkmb
|
phosphofructokinase, muscle b |
| chr21_+_7900107 | 0.56 |
ENSDART00000056560
|
ch25hl2
|
cholesterol 25-hydroxylase like 2 |
| chr20_-_26060154 | 0.56 |
ENSDART00000151950
|
serac1
|
serine active site containing 1 |
| chr8_-_15991473 | 0.55 |
ENSDART00000151746
|
agbl4
|
ATP/GTP binding protein-like 4 |
| chr8_-_15991801 | 0.55 |
ENSDART00000132005
|
agbl4
|
ATP/GTP binding protein-like 4 |
| chr7_-_31794476 | 0.55 |
ENSDART00000142385
|
nap1l4b
|
nucleosome assembly protein 1-like 4b |
| chr5_-_66301142 | 0.55 |
ENSDART00000067541
|
prlrb
|
prolactin receptor b |
| chr20_-_27711970 | 0.55 |
ENSDART00000139637
|
zbtb25
|
zinc finger and BTB domain containing 25 |
| chr15_+_29292154 | 0.55 |
ENSDART00000137817
|
rap1gap2a
|
RAP1 GTPase activating protein 2a |
| chr13_+_43050562 | 0.54 |
ENSDART00000016602
|
cdh23
|
cadherin-related 23 |
| chr8_+_25629941 | 0.54 |
ENSDART00000186925
|
wasa
|
Wiskott-Aldrich syndrome (eczema-thrombocytopenia) a |
| chr25_+_16214854 | 0.53 |
ENSDART00000109672
ENSDART00000190093 |
mical2b
|
microtubule associated monooxygenase, calponin and LIM domain containing 2b |
| chr17_-_47305034 | 0.53 |
ENSDART00000150116
|
alk
|
ALK receptor tyrosine kinase |
| chr6_+_40952031 | 0.53 |
ENSDART00000189219
|
patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr3_+_32553714 | 0.52 |
ENSDART00000165638
|
pax10
|
paired box 10 |
| chr7_+_65398161 | 0.51 |
ENSDART00000166109
ENSDART00000157399 |
usp47
|
ubiquitin specific peptidase 47 |
| chr6_-_24392426 | 0.50 |
ENSDART00000163965
|
brdt
|
bromodomain, testis-specific |
| chr18_+_27926839 | 0.49 |
ENSDART00000191835
|
hipk3b
|
homeodomain interacting protein kinase 3b |
| chr8_-_23578660 | 0.49 |
ENSDART00000039080
|
wasb
|
Wiskott-Aldrich syndrome (eczema-thrombocytopenia) b |
| chr1_+_44826367 | 0.49 |
ENSDART00000146962
|
STX3
|
zgc:165520 |
| chr19_+_34169055 | 0.48 |
ENSDART00000135592
ENSDART00000186043 |
poc1bl
|
POC1 centriolar protein homolog B (Chlamydomonas), like |
| chr17_+_450956 | 0.48 |
ENSDART00000183022
ENSDART00000171386 |
zgc:194887
|
zgc:194887 |
| chr22_-_11833317 | 0.48 |
ENSDART00000125423
ENSDART00000000192 |
ptpn4b
|
protein tyrosine phosphatase, non-receptor type 4b |
| chr8_+_51084993 | 0.48 |
ENSDART00000127709
ENSDART00000053768 |
DISP3
|
si:dkey-32e23.6 |
| chr15_+_40281693 | 0.48 |
ENSDART00000186565
|
gal3st2
|
galactose-3-O-sulfotransferase 2 |
| chr25_+_22017182 | 0.48 |
ENSDART00000156517
|
si:dkey-217l24.1
|
si:dkey-217l24.1 |
| chr19_+_2275019 | 0.47 |
ENSDART00000136138
|
itgb8
|
integrin, beta 8 |
| chr22_-_16997246 | 0.47 |
ENSDART00000090242
|
nfia
|
nuclear factor I/A |
| chr24_-_24848612 | 0.45 |
ENSDART00000190941
|
crhb
|
corticotropin releasing hormone b |
| chr16_-_50203058 | 0.45 |
ENSDART00000154570
|
vsig10l
|
V-set and immunoglobulin domain containing 10 like |
| chr8_+_53120278 | 0.45 |
ENSDART00000125232
|
nr2c2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr2_-_4787566 | 0.44 |
ENSDART00000160663
ENSDART00000157808 |
tnk2b
|
tyrosine kinase, non-receptor, 2b |
| chr7_+_34296789 | 0.43 |
ENSDART00000052471
ENSDART00000173798 ENSDART00000173778 |
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr8_-_45834825 | 0.42 |
ENSDART00000132965
|
ogdha
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase a (lipoamide) |
| chr5_-_50600512 | 0.42 |
ENSDART00000190318
|
mctp1a
|
multiple C2 domains, transmembrane 1a |
| chr2_-_44971551 | 0.41 |
ENSDART00000018818
|
mul1a
|
mitochondrial E3 ubiquitin protein ligase 1a |
| chr6_+_41957280 | 0.41 |
ENSDART00000050114
|
oxtr
|
oxytocin receptor |
| chr5_+_24282570 | 0.40 |
ENSDART00000041905
|
zdhhc23a
|
zinc finger, DHHC-type containing 23a |
| chr6_+_515181 | 0.38 |
ENSDART00000171374
|
CACNA1I (1 of many)
|
si:ch73-379f7.5 |
| chr13_+_22479988 | 0.38 |
ENSDART00000188182
ENSDART00000192972 ENSDART00000178372 |
ldb3a
|
LIM domain binding 3a |
| chr4_-_19921410 | 0.38 |
ENSDART00000184359
ENSDART00000179965 |
BX323820.1
|
|
| chr12_-_48127985 | 0.38 |
ENSDART00000193805
|
npffr1
|
neuropeptide FF receptor 1 |
| chr12_-_36268723 | 0.37 |
ENSDART00000113740
|
kcnj16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr4_-_2975461 | 0.37 |
ENSDART00000150794
|
plekha5
|
pleckstrin homology domain containing, family A member 5 |
| chr18_+_44769027 | 0.37 |
ENSDART00000145190
|
ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr5_+_10046643 | 0.36 |
ENSDART00000137543
ENSDART00000186917 |
gstt2
|
glutathione S-transferase theta 2 |
| chr3_-_41292275 | 0.35 |
ENSDART00000144088
|
sdk1a
|
sidekick cell adhesion molecule 1a |
| chr24_+_2961098 | 0.35 |
ENSDART00000163760
ENSDART00000170835 |
eci2
|
enoyl-CoA delta isomerase 2 |
| chr2_+_43755274 | 0.34 |
ENSDART00000138620
|
arhgap12a
|
Rho GTPase activating protein 12a |
| chr3_-_54500354 | 0.33 |
ENSDART00000124215
|
trip10a
|
thyroid hormone receptor interactor 10a |
| chr11_-_15090118 | 0.32 |
ENSDART00000171118
|
slc1a8a
|
solute carrier family 1 (glutamate transporter), member 8a |
| chr15_+_1705167 | 0.31 |
ENSDART00000081940
|
otol1b
|
otolin 1b |
| chr2_+_34767171 | 0.30 |
ENSDART00000145451
|
astn1
|
astrotactin 1 |
| chr17_+_30546579 | 0.30 |
ENSDART00000154385
|
nhsl1a
|
NHS-like 1a |
| chr16_-_45288599 | 0.30 |
ENSDART00000044097
|
hpn
|
hepsin |
| chr5_-_23200880 | 0.30 |
ENSDART00000051531
|
iqcd
|
IQ motif containing D |
| chr18_-_4957022 | 0.29 |
ENSDART00000101686
|
zbbx
|
zinc finger, B-box domain containing |
| chr3_-_22212764 | 0.29 |
ENSDART00000155490
|
maptb
|
microtubule-associated protein tau b |
| chr1_-_40208237 | 0.29 |
ENSDART00000191987
|
AL953865.1
|
|
| chr20_-_31743553 | 0.28 |
ENSDART00000087405
|
sash1a
|
SAM and SH3 domain containing 1a |
| chr14_-_17121676 | 0.27 |
ENSDART00000170154
ENSDART00000060479 |
smtnl1
|
smoothelin-like 1 |
| chr6_-_8498908 | 0.27 |
ENSDART00000149222
|
pglyrp2
|
peptidoglycan recognition protein 2 |
| chr11_+_5681762 | 0.26 |
ENSDART00000179139
|
arid3a
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr15_-_16946124 | 0.25 |
ENSDART00000154923
|
hip1
|
huntingtin interacting protein 1 |
| chr13_+_22480496 | 0.25 |
ENSDART00000136863
ENSDART00000131870 ENSDART00000078720 ENSDART00000078740 ENSDART00000139218 |
ldb3a
|
LIM domain binding 3a |
| chr15_-_21239416 | 0.24 |
ENSDART00000155787
|
A2ML1 (1 of many)
|
si:dkey-105h12.2 |
| chr8_-_979735 | 0.21 |
ENSDART00000149612
|
znf366
|
zinc finger protein 366 |
| chr3_-_51109286 | 0.20 |
ENSDART00000172010
|
si:ch211-148f13.1
|
si:ch211-148f13.1 |
| chr11_-_31226578 | 0.20 |
ENSDART00000109698
|
nacc1b
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing b |
| chr6_-_39765546 | 0.20 |
ENSDART00000185767
|
pfkmb
|
phosphofructokinase, muscle b |
| chr21_-_22724980 | 0.19 |
ENSDART00000035469
|
c1qa
|
complement component 1, q subcomponent, A chain |
| chr20_-_26937453 | 0.18 |
ENSDART00000139756
|
ftr97
|
finTRIM family, member 97 |
| chr7_-_31794878 | 0.17 |
ENSDART00000099748
|
nap1l4b
|
nucleosome assembly protein 1-like 4b |
| chr6_-_45995401 | 0.16 |
ENSDART00000154392
|
ca16b
|
carbonic anhydrase XVI b |
| chr7_-_44909540 | 0.15 |
ENSDART00000193378
ENSDART00000109534 |
cdh16
|
cadherin 16, KSP-cadherin |
| chr8_+_21406769 | 0.14 |
ENSDART00000135766
|
si:dkey-163f12.6
|
si:dkey-163f12.6 |
| chr3_+_32842825 | 0.13 |
ENSDART00000122228
|
prr14
|
proline rich 14 |
| chr23_+_26003187 | 0.12 |
ENSDART00000147271
|
wisp2
|
WNT1 inducible signaling pathway protein 2 |
| chr5_+_38462121 | 0.11 |
ENSDART00000144425
|
gltpd2
|
glycolipid transfer protein domain containing 2 |
| chr9_-_9419704 | 0.11 |
ENSDART00000138996
|
si:ch211-214p13.9
|
si:ch211-214p13.9 |
| chr20_-_26936887 | 0.10 |
ENSDART00000160827
|
ftr79
|
finTRIM family, member 79 |
| chr17_+_30545895 | 0.09 |
ENSDART00000076739
|
nhsl1a
|
NHS-like 1a |
| chr20_-_48701593 | 0.09 |
ENSDART00000132835
|
pax1b
|
paired box 1b |
| chr22_+_24673168 | 0.05 |
ENSDART00000135257
|
si:rp71-23d18.8
|
si:rp71-23d18.8 |
| chr14_-_32258759 | 0.05 |
ENSDART00000052949
|
fgf13a
|
fibroblast growth factor 13a |
| chr18_+_9382847 | 0.05 |
ENSDART00000061886
|
sema3ab
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Ab |
| chr20_-_25369767 | 0.03 |
ENSDART00000180278
|
itsn2a
|
intersectin 2a |
| chr5_+_19413838 | 0.02 |
ENSDART00000089014
ENSDART00000146215 |
myo1ha
|
myosin IHa |
| chr13_-_42673978 | 0.01 |
ENSDART00000133848
ENSDART00000099738 ENSDART00000099729 ENSDART00000169083 |
lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr4a1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.5 | 3.3 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.4 | 1.7 | GO:0006867 | asparagine transport(GO:0006867) |
| 0.4 | 2.0 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.3 | 1.0 | GO:1903961 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.3 | 1.7 | GO:0097066 | response to thyroid hormone(GO:0097066) |
| 0.3 | 1.6 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.3 | 1.4 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.3 | 4.9 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.3 | 2.8 | GO:0097090 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.2 | 0.5 | GO:0051037 | histone displacement(GO:0001207) regulation of transcription involved in meiotic cell cycle(GO:0051037) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.2 | 1.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.2 | 0.5 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.1 | 1.6 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.5 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 0.4 | GO:0007567 | parturition(GO:0007567) neurohypophysis development(GO:0021985) maternal process involved in parturition(GO:0060137) |
| 0.1 | 2.0 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.1 | 0.9 | GO:0043090 | amino acid import(GO:0043090) L-amino acid import(GO:0043092) |
| 0.1 | 1.1 | GO:0071450 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.1 | 0.5 | GO:0035678 | neuromast hair cell morphogenesis(GO:0035678) |
| 0.1 | 3.0 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.1 | 0.3 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.1 | 0.7 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.1 | 0.4 | GO:0006549 | isoleucine metabolic process(GO:0006549) isoleucine biosynthetic process(GO:0009097) |
| 0.1 | 1.2 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 0.5 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 1.0 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 0.9 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 1.1 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.1 | 1.2 | GO:0010962 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.1 | 0.8 | GO:0061615 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 1.0 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 4.7 | GO:0032869 | cellular response to insulin stimulus(GO:0032869) |
| 0.1 | 1.8 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.9 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 0.3 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 2.1 | GO:0005977 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.0 | 1.5 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 1.0 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 1.1 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.1 | GO:0016045 | detection of biotic stimulus(GO:0009595) detection of bacterium(GO:0016045) detection of other organism(GO:0098543) detection of external biotic stimulus(GO:0098581) |
| 0.0 | 1.4 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.4 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.4 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.0 | 3.0 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.9 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.9 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 4.0 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 0.5 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 0.6 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 0.6 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.4 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.6 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.5 | GO:0031647 | regulation of protein stability(GO:0031647) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.0 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.3 | 4.9 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.2 | 2.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 1.7 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 1.5 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 0.4 | GO:0005948 | acetolactate synthase complex(GO:0005948) |
| 0.1 | 0.3 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.1 | 1.6 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 1.8 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.1 | 1.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.8 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.3 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.0 | 2.0 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.8 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.9 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 3.0 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 1.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.6 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.9 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.9 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.5 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.5 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.0 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.5 | 1.4 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.4 | 2.7 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.4 | 1.2 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.3 | 1.6 | GO:0051430 | mu-type opioid receptor binding(GO:0031852) dopamine receptor binding(GO:0050780) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.3 | 1.7 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.3 | 1.8 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.2 | 0.8 | GO:0015105 | arsenite transmembrane transporter activity(GO:0015105) |
| 0.2 | 1.5 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.2 | 1.8 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 1.6 | GO:0015386 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 1.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 0.5 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.1 | 0.4 | GO:0004990 | oxytocin receptor activity(GO:0004990) |
| 0.1 | 0.4 | GO:0003984 | acetolactate synthase activity(GO:0003984) |
| 0.1 | 3.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.7 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 1.0 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 1.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 1.1 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 0.5 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.1 | 0.6 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.1 | 1.2 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 1.8 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 2.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.8 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 0.9 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 0.5 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.4 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.5 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.3 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.6 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.1 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.0 | 0.9 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.9 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.8 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.3 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 3.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 2.0 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.9 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 3.3 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.6 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.4 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 2.0 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 4.7 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 2.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.3 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.6 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 2.1 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.4 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.4 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 3.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.5 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.8 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 1.0 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.6 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.4 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.0 | PID P73PATHWAY | p73 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.1 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 2.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 3.6 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 1.6 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.1 | 0.7 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 0.6 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 1.0 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 2.0 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 0.9 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 3.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 0.8 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 0.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 1.6 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.8 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 1.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.8 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |