Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
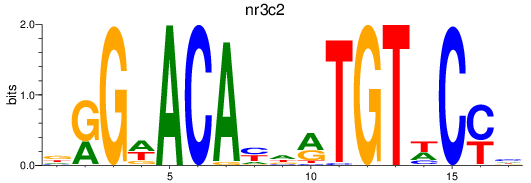
Results for nr3c2
Z-value: 0.59
Transcription factors associated with nr3c2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr3c2
|
ENSDARG00000102082 | nuclear receptor subfamily 3, group C, member 2 |
|
nr3c2
|
ENSDARG00000115513 | nuclear receptor subfamily 3, group C, member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr3c2 | dr11_v1_chr1_-_37087966_37087966 | 0.01 | 9.8e-01 | Click! |
Activity profile of nr3c2 motif
Sorted Z-values of nr3c2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_41096058 | 2.16 |
ENSDART00000028373
|
fkbp5
|
FK506 binding protein 5 |
| chr6_+_41099787 | 1.28 |
ENSDART00000186884
|
fkbp5
|
FK506 binding protein 5 |
| chr18_-_24996634 | 0.99 |
ENSDART00000170210
|
si:ch211-196l7.4
|
si:ch211-196l7.4 |
| chr15_+_37559570 | 0.85 |
ENSDART00000085522
|
hspb6
|
heat shock protein, alpha-crystallin-related, b6 |
| chr24_-_38097305 | 0.84 |
ENSDART00000124321
|
crp2
|
C-reactive protein 2 |
| chr10_+_33754967 | 0.62 |
ENSDART00000153442
|
rxfp2a
|
relaxin/insulin-like family peptide receptor 2a |
| chr15_+_40188076 | 0.56 |
ENSDART00000063779
|
efhd1
|
EF-hand domain family, member D1 |
| chr16_+_50089417 | 0.56 |
ENSDART00000153675
|
nr1d2a
|
nuclear receptor subfamily 1, group D, member 2a |
| chr15_-_24883956 | 0.55 |
ENSDART00000113199
|
aipl1
|
aryl hydrocarbon receptor interacting protein-like 1 |
| chr7_+_48288762 | 0.49 |
ENSDART00000083569
|
oaz2b
|
ornithine decarboxylase antizyme 2b |
| chr7_+_58699718 | 0.46 |
ENSDART00000049264
|
sdr16c5b
|
short chain dehydrogenase/reductase family 16C, member 5b |
| chr10_-_24689725 | 0.42 |
ENSDART00000079566
|
si:ch211-287a12.9
|
si:ch211-287a12.9 |
| chr10_+_26667475 | 0.38 |
ENSDART00000133281
ENSDART00000147013 |
si:ch73-52f15.5
|
si:ch73-52f15.5 |
| chr17_+_6276559 | 0.35 |
ENSDART00000131075
|
dusp23b
|
dual specificity phosphatase 23b |
| chr6_+_50451337 | 0.33 |
ENSDART00000155051
|
mych
|
myelocytomatosis oncogene homolog |
| chr9_+_22388686 | 0.32 |
ENSDART00000182731
ENSDART00000181462 |
dgkg
|
diacylglycerol kinase, gamma |
| chr5_+_9382301 | 0.32 |
ENSDART00000124017
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr11_+_5880562 | 0.32 |
ENSDART00000129663
ENSDART00000130768 ENSDART00000160909 |
dazap1
|
DAZ associated protein 1 |
| chr20_-_38836161 | 0.31 |
ENSDART00000061358
|
si:dkey-221h15.4
|
si:dkey-221h15.4 |
| chr5_+_9417409 | 0.29 |
ENSDART00000125421
ENSDART00000130265 |
ugt2b5
|
UDP glucuronosyltransferase 2 family, polypeptide B5 |
| chr11_+_34522554 | 0.25 |
ENSDART00000109833
|
zmat3
|
zinc finger, matrin-type 3 |
| chr10_-_13178853 | 0.24 |
ENSDART00000163740
ENSDART00000166327 ENSDART00000160265 ENSDART00000164299 |
musk
|
muscle, skeletal, receptor tyrosine kinase |
| chr2_+_29257942 | 0.24 |
ENSDART00000184362
ENSDART00000025562 |
cdh18a
|
cadherin 18, type 2a |
| chr23_+_44732863 | 0.24 |
ENSDART00000160044
ENSDART00000172268 |
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr14_-_28001986 | 0.23 |
ENSDART00000054115
|
tsc22d3
|
TSC22 domain family, member 3 |
| chr11_-_34147205 | 0.23 |
ENSDART00000173216
|
atp13a3
|
ATPase 13A3 |
| chr2_-_48396061 | 0.22 |
ENSDART00000161657
|
si:ch211-195j11.30
|
si:ch211-195j11.30 |
| chr9_+_23895711 | 0.20 |
ENSDART00000034686
|
cops8
|
COP9 signalosome subunit 8 |
| chr18_-_19491162 | 0.20 |
ENSDART00000090413
|
snapc5
|
small nuclear RNA activating complex, polypeptide 5 |
| chr25_-_3470910 | 0.20 |
ENSDART00000029067
ENSDART00000186737 |
hbp1
|
HMG-box transcription factor 1 |
| chr5_+_23256187 | 0.17 |
ENSDART00000168717
ENSDART00000142915 |
si:dkey-125i10.3
|
si:dkey-125i10.3 |
| chr17_+_29345606 | 0.16 |
ENSDART00000086164
|
kctd3
|
potassium channel tetramerization domain containing 3 |
| chr5_+_9360394 | 0.16 |
ENSDART00000124642
|
FP236810.2
|
|
| chr21_+_6291027 | 0.16 |
ENSDART00000180467
ENSDART00000184952 ENSDART00000184006 |
fnbp1b
|
formin binding protein 1b |
| chr5_+_30518441 | 0.15 |
ENSDART00000132664
|
hmbsa
|
hydroxymethylbilane synthase a |
| chr21_+_6290566 | 0.14 |
ENSDART00000161647
|
fnbp1b
|
formin binding protein 1b |
| chr22_-_36519590 | 0.14 |
ENSDART00000129318
|
CABZ01045212.1
|
|
| chr23_-_16734009 | 0.14 |
ENSDART00000125449
|
si:ch211-224l10.4
|
si:ch211-224l10.4 |
| chr25_+_10416583 | 0.13 |
ENSDART00000073907
|
ehf
|
ets homologous factor |
| chr14_+_52571134 | 0.13 |
ENSDART00000166708
|
rpl26
|
ribosomal protein L26 |
| chr11_+_34523132 | 0.12 |
ENSDART00000192257
|
zmat3
|
zinc finger, matrin-type 3 |
| chr9_-_30145080 | 0.12 |
ENSDART00000133746
|
abi3bpa
|
ABI family, member 3 (NESH) binding protein a |
| chr20_-_39391833 | 0.11 |
ENSDART00000135149
|
si:dkey-217m5.8
|
si:dkey-217m5.8 |
| chr16_+_46430627 | 0.09 |
ENSDART00000127681
|
rpz6
|
rapunzel 6 |
| chr24_-_23784701 | 0.07 |
ENSDART00000090368
|
sgk3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr18_-_20608300 | 0.05 |
ENSDART00000140632
|
bcl2l13
|
BCL2 like 13 |
| chr16_-_11932923 | 0.04 |
ENSDART00000103975
|
cd4-1
|
CD4-1 molecule |
| chr6_-_27057702 | 0.04 |
ENSDART00000149363
|
stk25a
|
serine/threonine kinase 25a |
| chr10_+_20364009 | 0.04 |
ENSDART00000186139
ENSDART00000080395 |
golga7
|
golgin A7 |
| chr17_-_6613458 | 0.04 |
ENSDART00000175024
|
si:ch211-189e2.3
|
si:ch211-189e2.3 |
| chr15_+_31462664 | 0.03 |
ENSDART00000174267
ENSDART00000185665 ENSDART00000060087 |
or102-1
|
odorant receptor, family C, subfamily 102, member 1 |
| chr3_-_37476475 | 0.03 |
ENSDART00000148107
|
si:ch211-278a6.1
|
si:ch211-278a6.1 |
| chr10_-_24689280 | 0.03 |
ENSDART00000191476
|
si:ch211-287a12.9
|
si:ch211-287a12.9 |
| chr22_-_16275236 | 0.02 |
ENSDART00000149051
|
cdc14ab
|
cell division cycle 14Ab |
| chr24_-_20641000 | 0.02 |
ENSDART00000166135
|
zbtb47b
|
zinc finger and BTB domain containing 47b |
| chr6_+_4255319 | 0.01 |
ENSDART00000170351
|
nbeal1
|
neurobeachin-like 1 |
| chr22_+_19538626 | 0.00 |
ENSDART00000189174
|
BX936298.2
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of nr3c2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.2 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.6 | GO:0045762 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.0 | 0.2 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.2 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 0.2 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.0 | 0.5 | GO:0006595 | polyamine metabolic process(GO:0006595) |
| 0.0 | 0.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.3 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.2 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.6 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.8 | GO:0005811 | lipid particle(GO:0005811) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.4 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.5 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.3 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.2 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.0 | 0.8 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 3.4 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |