Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
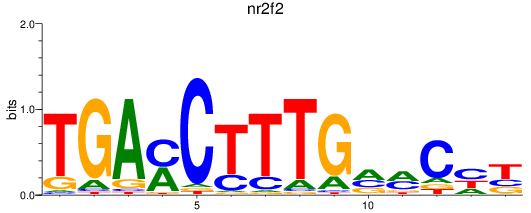
Results for nr2f2
Z-value: 0.89
Transcription factors associated with nr2f2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr2f2
|
ENSDARG00000040926 | nuclear receptor subfamily 2, group F, member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr2f2 | dr11_v1_chr18_-_23874929_23874929 | 0.42 | 7.4e-02 | Click! |
Activity profile of nr2f2 motif
Sorted Z-values of nr2f2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_45128375 | 2.45 |
ENSDART00000164805
|
camk2b2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 2 |
| chr17_-_25737452 | 2.13 |
ENSDART00000152021
|
si:ch211-214p16.3
|
si:ch211-214p16.3 |
| chr24_-_29586082 | 1.91 |
ENSDART00000136763
|
vav3a
|
vav 3 guanine nucleotide exchange factor a |
| chr17_-_14613711 | 1.71 |
ENSDART00000157345
|
sdsl
|
serine dehydratase-like |
| chr1_+_25848231 | 1.65 |
ENSDART00000027973
|
fabp2
|
fatty acid binding protein 2, intestinal |
| chr4_-_16451375 | 1.61 |
ENSDART00000192700
ENSDART00000128835 |
wu:fc23c09
|
wu:fc23c09 |
| chr4_-_5317006 | 1.53 |
ENSDART00000150867
|
si:ch211-214j24.15
|
si:ch211-214j24.15 |
| chr10_-_11385155 | 1.50 |
ENSDART00000064214
|
plac8.1
|
placenta-specific 8, tandem duplicate 1 |
| chr7_-_26457208 | 1.44 |
ENSDART00000173519
|
zgc:172079
|
zgc:172079 |
| chr12_+_22607761 | 1.41 |
ENSDART00000153112
|
si:dkey-219e21.2
|
si:dkey-219e21.2 |
| chr9_-_46382637 | 1.38 |
ENSDART00000085738
|
lct
|
lactase |
| chr7_-_5487593 | 1.35 |
ENSDART00000136594
|
arhgef11
|
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr24_-_14712427 | 1.32 |
ENSDART00000176316
|
jph1a
|
junctophilin 1a |
| chr22_+_35089031 | 1.31 |
ENSDART00000076040
|
srfa
|
serum response factor a |
| chr7_+_73444325 | 1.30 |
ENSDART00000123016
|
si:ch211-142d6.2
|
si:ch211-142d6.2 |
| chr22_-_31517300 | 1.20 |
ENSDART00000164799
|
slc6a6b
|
solute carrier family 6 (neurotransmitter transporter), member 6b |
| chr20_-_25522911 | 1.14 |
ENSDART00000063058
|
cyp2n13
|
cytochrome P450, family 2, subfamily N, polypeptide 13 |
| chr17_+_450956 | 1.11 |
ENSDART00000183022
ENSDART00000171386 |
zgc:194887
|
zgc:194887 |
| chr12_-_31457301 | 1.09 |
ENSDART00000043887
ENSDART00000148603 |
acsl5
|
acyl-CoA synthetase long chain family member 5 |
| chr9_-_9982696 | 1.05 |
ENSDART00000192548
ENSDART00000125852 |
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr2_-_8102169 | 1.04 |
ENSDART00000131955
|
pls1
|
plastin 1 (I isoform) |
| chr5_+_8919698 | 1.00 |
ENSDART00000046440
|
agpat9l
|
1-acylglycerol-3-phosphate O-acyltransferase 9, like |
| chr19_-_677713 | 0.96 |
ENSDART00000025146
|
slc6a19a.1
|
solute carrier family 6 (neutral amino acid transporter), member 19a, tandem duplicate 1 |
| chr22_+_15331214 | 0.94 |
ENSDART00000136566
|
sult3st4
|
sulfotransferase family 3, cytosolic sulfotransferase 4 |
| chr25_-_12803723 | 0.93 |
ENSDART00000158787
|
ca5a
|
carbonic anhydrase Va |
| chr20_+_25568694 | 0.92 |
ENSDART00000063107
ENSDART00000063128 |
cyp2p7
|
cytochrome P450, family 2, subfamily P, polypeptide 7 |
| chr8_+_43053519 | 0.91 |
ENSDART00000147178
|
prnpa
|
prion protein a |
| chr5_-_36549024 | 0.90 |
ENSDART00000097671
|
zgc:158432
|
zgc:158432 |
| chr23_-_44786844 | 0.89 |
ENSDART00000148669
|
si:ch73-269m23.5
|
si:ch73-269m23.5 |
| chr4_+_2655358 | 0.88 |
ENSDART00000007638
|
bcap29
|
B cell receptor associated protein 29 |
| chr9_+_30633184 | 0.87 |
ENSDART00000191310
|
tbc1d4
|
TBC1 domain family, member 4 |
| chr5_-_3839285 | 0.87 |
ENSDART00000122292
|
mlxipl
|
MLX interacting protein like |
| chr17_+_13031497 | 0.86 |
ENSDART00000115208
|
fbxo33
|
F-box protein 33 |
| chr3_+_12744083 | 0.85 |
ENSDART00000158554
ENSDART00000169545 |
cyp2k21
|
cytochrome P450, family 2, subfamily k, polypeptide 21 |
| chr23_-_18030399 | 0.84 |
ENSDART00000136967
|
pm20d1.1
|
peptidase M20 domain containing 1, tandem duplicate 1 |
| chr20_-_15090862 | 0.83 |
ENSDART00000063892
ENSDART00000122592 |
si:dkey-239i20.2
|
si:dkey-239i20.2 |
| chr4_-_31064105 | 0.83 |
ENSDART00000157670
|
si:dkey-11n14.1
|
si:dkey-11n14.1 |
| chr7_+_65398161 | 0.82 |
ENSDART00000166109
ENSDART00000157399 |
usp47
|
ubiquitin specific peptidase 47 |
| chr15_-_1843831 | 0.81 |
ENSDART00000156718
ENSDART00000154175 |
taf15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr3_+_12732382 | 0.81 |
ENSDART00000158403
|
cyp2k19
|
cytochrome P450, family 2, subfamily k, polypeptide 19 |
| chr11_-_6974022 | 0.81 |
ENSDART00000172851
|
COMP
|
si:ch211-43f4.1 |
| chr18_+_26829086 | 0.79 |
ENSDART00000098356
|
slc28a1
|
solute carrier family 28 (concentrative nucleoside transporter), member 1 |
| chr12_+_26670778 | 0.78 |
ENSDART00000144355
|
arhgap12b
|
Rho GTPase activating protein 12b |
| chr3_-_46410387 | 0.77 |
ENSDART00000156822
|
cdip1
|
cell death-inducing p53 target 1 |
| chr4_-_8152746 | 0.77 |
ENSDART00000012928
ENSDART00000177482 |
wnk1b
|
WNK lysine deficient protein kinase 1b |
| chr13_-_3516473 | 0.77 |
ENSDART00000146240
|
prkn
|
parkin RBR E3 ubiquitin protein ligase |
| chr9_+_9502610 | 0.76 |
ENSDART00000061525
ENSDART00000125174 |
nr1i2
|
nuclear receptor subfamily 1, group I, member 2 |
| chr18_-_12957451 | 0.75 |
ENSDART00000140403
|
srgap1a
|
SLIT-ROBO Rho GTPase activating protein 1a |
| chr19_+_3056450 | 0.75 |
ENSDART00000141324
ENSDART00000082353 |
hsf1
|
heat shock transcription factor 1 |
| chr3_-_16227683 | 0.74 |
ENSDART00000111707
|
cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr17_-_29194219 | 0.74 |
ENSDART00000157340
|
sptbn5
|
spectrin, beta, non-erythrocytic 5 |
| chr8_+_47188154 | 0.74 |
ENSDART00000137319
|
si:dkeyp-100a1.6
|
si:dkeyp-100a1.6 |
| chr18_+_36037223 | 0.74 |
ENSDART00000144410
|
tmem91
|
transmembrane protein 91 |
| chr2_-_53592532 | 0.73 |
ENSDART00000184066
|
ccl25a
|
chemokine (C-C motif) ligand 25a |
| chr12_+_49125510 | 0.73 |
ENSDART00000185804
|
FO704607.1
|
|
| chr25_-_21085661 | 0.72 |
ENSDART00000099355
|
prr5a
|
proline rich 5a (renal) |
| chr20_+_43379029 | 0.71 |
ENSDART00000142486
ENSDART00000186486 |
unc93a
|
unc-93 homolog A |
| chr3_-_18805225 | 0.71 |
ENSDART00000133471
ENSDART00000131758 |
msrb1a
|
methionine sulfoxide reductase B1a |
| chr8_+_999421 | 0.71 |
ENSDART00000149528
|
fabp1b.1
|
fatty acid binding protein 1b, tandem duplicate 1 |
| chr6_-_49873020 | 0.70 |
ENSDART00000148511
|
gnas
|
GNAS complex locus |
| chr24_-_1341543 | 0.70 |
ENSDART00000169341
|
nrp1a
|
neuropilin 1a |
| chr25_-_31118923 | 0.70 |
ENSDART00000009126
ENSDART00000188286 |
kras
|
v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog |
| chr18_+_27926839 | 0.69 |
ENSDART00000191835
|
hipk3b
|
homeodomain interacting protein kinase 3b |
| chr3_-_4455951 | 0.69 |
ENSDART00000193908
ENSDART00000074077 |
trim35-3
|
tripartite motif containing 35-3 |
| chr2_+_43755274 | 0.68 |
ENSDART00000138620
|
arhgap12a
|
Rho GTPase activating protein 12a |
| chr4_-_858434 | 0.68 |
ENSDART00000006961
|
sobpb
|
sine oculis binding protein homolog (Drosophila) b |
| chr25_-_19420949 | 0.67 |
ENSDART00000181338
|
map1ab
|
microtubule-associated protein 1Ab |
| chr17_+_12408405 | 0.66 |
ENSDART00000154827
ENSDART00000048440 ENSDART00000156429 |
khk
|
ketohexokinase |
| chr11_+_30314885 | 0.66 |
ENSDART00000187418
ENSDART00000123244 |
ugt1b2
|
UDP glucuronosyltransferase 1 family, polypeptide B2 |
| chr23_-_45504991 | 0.66 |
ENSDART00000148761
|
col24a1
|
collagen type XXIV alpha 1 |
| chr23_-_46126444 | 0.65 |
ENSDART00000030004
|
CABZ01071903.1
|
|
| chr2_-_57076687 | 0.63 |
ENSDART00000161523
|
slc25a42
|
solute carrier family 25, member 42 |
| chr8_-_33154677 | 0.63 |
ENSDART00000133300
|
zbtb34
|
zinc finger and BTB domain containing 34 |
| chr14_+_740032 | 0.63 |
ENSDART00000165014
|
klb
|
klotho beta |
| chr12_-_20362041 | 0.62 |
ENSDART00000184145
ENSDART00000105952 |
aqp8a.2
|
aquaporin 8a, tandem duplicate 2 |
| chr7_-_58244220 | 0.61 |
ENSDART00000180450
|
unm_hu7910
|
un-named hu7910 |
| chr15_+_47618221 | 0.61 |
ENSDART00000168722
|
paf1
|
PAF1 homolog, Paf1/RNA polymerase II complex component |
| chr18_+_26829362 | 0.61 |
ENSDART00000132728
|
slc28a1
|
solute carrier family 28 (concentrative nucleoside transporter), member 1 |
| chr4_-_18309917 | 0.60 |
ENSDART00000189084
|
plxnc1
|
plexin C1 |
| chr21_-_9446747 | 0.60 |
ENSDART00000158790
|
ptpn13
|
protein tyrosine phosphatase, non-receptor type 13 |
| chr15_-_34418525 | 0.60 |
ENSDART00000147582
|
agmo
|
alkylglycerol monooxygenase |
| chr15_-_30857350 | 0.59 |
ENSDART00000138988
|
akap1b
|
A kinase (PRKA) anchor protein 1b |
| chr21_-_41617372 | 0.59 |
ENSDART00000187171
|
BX005397.1
|
|
| chr25_-_31898552 | 0.59 |
ENSDART00000156128
|
si:ch73-330k17.3
|
si:ch73-330k17.3 |
| chr23_+_44633858 | 0.59 |
ENSDART00000180728
|
si:ch73-265d7.2
|
si:ch73-265d7.2 |
| chr20_+_36682051 | 0.58 |
ENSDART00000130513
|
ncoa1
|
nuclear receptor coactivator 1 |
| chr21_-_21096437 | 0.57 |
ENSDART00000186552
|
ank1b
|
ankyrin 1, erythrocytic b |
| chr24_-_11127493 | 0.56 |
ENSDART00000144066
|
myrip
|
myosin VIIA and Rab interacting protein |
| chr7_-_24520866 | 0.56 |
ENSDART00000077039
|
faah2b
|
fatty acid amide hydrolase 2b |
| chr22_-_16400484 | 0.56 |
ENSDART00000135987
|
lama3
|
laminin, alpha 3 |
| chr24_+_39137001 | 0.56 |
ENSDART00000181086
ENSDART00000183724 ENSDART00000193466 |
tbc1d24
|
TBC1 domain family, member 24 |
| chr11_+_6116096 | 0.55 |
ENSDART00000159680
|
nr2f6b
|
nuclear receptor subfamily 2, group F, member 6b |
| chr6_+_39499623 | 0.55 |
ENSDART00000036057
|
si:ch211-173n18.3
|
si:ch211-173n18.3 |
| chr1_-_58868306 | 0.55 |
ENSDART00000166615
|
dnm2b
|
dynamin 2b |
| chr4_-_53354589 | 0.54 |
ENSDART00000169031
|
si:dkey-250k10.4
|
si:dkey-250k10.4 |
| chr16_+_4078240 | 0.53 |
ENSDART00000160890
|
inpp5b
|
inositol polyphosphate-5-phosphatase B |
| chr1_-_19215336 | 0.53 |
ENSDART00000162949
ENSDART00000170680 |
ptprdb
|
protein tyrosine phosphatase, receptor type, D, b |
| chr3_+_1179601 | 0.52 |
ENSDART00000173378
|
triobpb
|
TRIO and F-actin binding protein b |
| chr3_-_1317290 | 0.52 |
ENSDART00000047094
|
LO018552.1
|
|
| chr15_-_2726662 | 0.51 |
ENSDART00000108856
|
si:ch211-235m3.5
|
si:ch211-235m3.5 |
| chr13_-_9886579 | 0.51 |
ENSDART00000101926
|
si:ch211-117n7.7
|
si:ch211-117n7.7 |
| chr7_-_38183331 | 0.51 |
ENSDART00000149382
|
abcc12
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 12 |
| chr23_-_18017946 | 0.50 |
ENSDART00000104592
|
pm20d1.2
|
peptidase M20 domain containing 1, tandem duplicate 2 |
| chr8_-_53108207 | 0.50 |
ENSDART00000111023
|
b3galt4
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 4 |
| chr3_+_30257582 | 0.50 |
ENSDART00000159497
ENSDART00000103457 ENSDART00000121883 |
mybpc2a
|
myosin binding protein C, fast type a |
| chr2_-_42960353 | 0.50 |
ENSDART00000098303
|
oc90
|
otoconin 90 |
| chr25_+_34246625 | 0.50 |
ENSDART00000082578
|
bnip2
|
BCL2 interacting protein 2 |
| chr4_+_25917915 | 0.50 |
ENSDART00000138603
|
vezt
|
vezatin, adherens junctions transmembrane protein |
| chr12_-_36260532 | 0.49 |
ENSDART00000022533
|
kcnj2a
|
potassium inwardly-rectifying channel, subfamily J, member 2a |
| chr20_+_2281933 | 0.49 |
ENSDART00000137579
|
si:ch73-18b11.2
|
si:ch73-18b11.2 |
| chr25_+_418932 | 0.48 |
ENSDART00000059193
|
prtgb
|
protogenin homolog b (Gallus gallus) |
| chr4_-_19693978 | 0.48 |
ENSDART00000100974
ENSDART00000040405 |
snd1
|
staphylococcal nuclease and tudor domain containing 1 |
| chr15_+_28268135 | 0.48 |
ENSDART00000152536
ENSDART00000188550 |
myo1cb
|
myosin Ic, paralog b |
| chr20_+_50115335 | 0.48 |
ENSDART00000031139
|
slc24a4b
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4b |
| chr15_+_47386939 | 0.47 |
ENSDART00000128224
|
FO904873.1
|
|
| chr25_+_22017182 | 0.47 |
ENSDART00000156517
|
si:dkey-217l24.1
|
si:dkey-217l24.1 |
| chr13_-_37254777 | 0.47 |
ENSDART00000139734
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr16_-_22294265 | 0.47 |
ENSDART00000124718
|
aqp10a
|
aquaporin 10a |
| chr9_+_38075851 | 0.46 |
ENSDART00000135314
|
cacnb4a
|
calcium channel, voltage-dependent, beta 4a subunit |
| chr5_+_31605322 | 0.46 |
ENSDART00000135037
|
si:dkey-220k22.3
|
si:dkey-220k22.3 |
| chr22_-_3595439 | 0.46 |
ENSDART00000083308
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr24_-_26715174 | 0.45 |
ENSDART00000079726
|
pld1b
|
phospholipase D1b |
| chr17_-_15274648 | 0.45 |
ENSDART00000141257
|
ros1
|
c-ros oncogene 1 , receptor tyrosine kinase |
| chr5_+_43470544 | 0.45 |
ENSDART00000111587
|
rhobtb2a
|
Rho-related BTB domain containing 2a |
| chr3_+_12718100 | 0.44 |
ENSDART00000162343
ENSDART00000192425 |
cyp2k20
|
cytochrome P450, family 2, subfamily k, polypeptide 20 |
| chr5_-_67115872 | 0.44 |
ENSDART00000065262
|
rps6ka4
|
ribosomal protein S6 kinase, polypeptide 4 |
| chr8_+_36339470 | 0.44 |
ENSDART00000191250
|
FO744840.1
|
|
| chr9_+_32859967 | 0.43 |
ENSDART00000168992
|
si:dkey-145p14.5
|
si:dkey-145p14.5 |
| chr1_+_58094551 | 0.42 |
ENSDART00000146316
|
si:ch211-114l13.1
|
si:ch211-114l13.1 |
| chr5_-_24124118 | 0.42 |
ENSDART00000051550
|
capga
|
capping protein (actin filament), gelsolin-like a |
| chr5_-_33607062 | 0.42 |
ENSDART00000048350
|
si:dkey-34e4.1
|
si:dkey-34e4.1 |
| chr20_-_52967878 | 0.42 |
ENSDART00000164460
|
gata4
|
GATA binding protein 4 |
| chr13_-_41546779 | 0.42 |
ENSDART00000163331
|
pcdh15a
|
protocadherin-related 15a |
| chr18_+_5172848 | 0.41 |
ENSDART00000190642
|
CABZ01080599.1
|
|
| chr2_+_3533458 | 0.41 |
ENSDART00000133007
|
gpt2l
|
glutamic pyruvate transaminase (alanine aminotransferase) 2, like |
| chr25_-_18002937 | 0.40 |
ENSDART00000149696
|
cep290
|
centrosomal protein 290 |
| chr17_+_25856671 | 0.39 |
ENSDART00000064817
|
wapla
|
WAPL cohesin release factor a |
| chr25_-_35143360 | 0.39 |
ENSDART00000188033
|
zgc:165555
|
zgc:165555 |
| chr20_+_39250673 | 0.39 |
ENSDART00000153003
|
reps1
|
RALBP1 associated Eps domain containing 1 |
| chr19_-_27564980 | 0.39 |
ENSDART00000171967
|
si:dkeyp-46h3.8
|
si:dkeyp-46h3.8 |
| chr21_+_34992550 | 0.38 |
ENSDART00000109041
ENSDART00000135400 |
tmprss15
|
transmembrane protease, serine 15 |
| chr25_+_34247353 | 0.38 |
ENSDART00000148914
|
bnip2
|
BCL2 interacting protein 2 |
| chr20_+_46213553 | 0.38 |
ENSDART00000100532
|
stx7l
|
syntaxin 7-like |
| chr3_+_12755535 | 0.38 |
ENSDART00000161286
|
cyp2k17
|
cytochrome P450, family 2, subfamily K, polypeptide17 |
| chr4_-_9533345 | 0.38 |
ENSDART00000128501
|
si:ch73-139j3.4
|
si:ch73-139j3.4 |
| chr6_-_55354004 | 0.38 |
ENSDART00000165911
|
pcif1
|
PDX1 C-terminal inhibiting factor 1 |
| chr18_-_37007294 | 0.38 |
ENSDART00000088309
|
map3k10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr22_+_15323930 | 0.38 |
ENSDART00000142416
|
si:dkey-236e20.3
|
si:dkey-236e20.3 |
| chr7_+_38267136 | 0.38 |
ENSDART00000173613
|
gpatch1
|
G patch domain containing 1 |
| chr7_+_59649399 | 0.37 |
ENSDART00000123520
ENSDART00000040771 |
rpl34
|
ribosomal protein L34 |
| chr11_+_33284837 | 0.37 |
ENSDART00000166239
ENSDART00000111412 |
nkpd1
|
NTPase, KAP family P-loop domain containing 1 |
| chr5_+_32009080 | 0.37 |
ENSDART00000186885
|
scai
|
suppressor of cancer cell invasion |
| chr4_-_47096170 | 0.37 |
ENSDART00000161079
|
si:dkey-26m3.3
|
si:dkey-26m3.3 |
| chr21_-_28523548 | 0.37 |
ENSDART00000077910
|
epdl2
|
ependymin-like 2 |
| chr4_+_33547662 | 0.37 |
ENSDART00000150439
|
si:dkey-84h14.2
|
si:dkey-84h14.2 |
| chr19_-_3056235 | 0.36 |
ENSDART00000137020
|
bop1
|
block of proliferation 1 |
| chr4_-_9579299 | 0.36 |
ENSDART00000183079
ENSDART00000192968 ENSDART00000091809 |
shank3b
|
SH3 and multiple ankyrin repeat domains 3b |
| chr10_-_21953643 | 0.36 |
ENSDART00000188921
ENSDART00000193569 |
FO744833.2
|
|
| chr17_-_6076266 | 0.35 |
ENSDART00000171084
|
ephx2
|
epoxide hydrolase 2, cytoplasmic |
| chr8_-_19051906 | 0.35 |
ENSDART00000089024
|
sema6bb
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6Bb |
| chr2_+_42715995 | 0.35 |
ENSDART00000143419
ENSDART00000183914 ENSDART00000184371 ENSDART00000185485 ENSDART00000037332 |
ftr12
|
finTRIM family, member 12 |
| chr5_-_7897316 | 0.35 |
ENSDART00000160743
ENSDART00000160912 |
opn4xb
|
opsin 4xb |
| chr1_-_44314 | 0.34 |
ENSDART00000160050
|
tmem39a
|
transmembrane protein 39A |
| chr9_+_17984358 | 0.34 |
ENSDART00000192399
|
akap11
|
A kinase (PRKA) anchor protein 11 |
| chr2_-_20120904 | 0.34 |
ENSDART00000186002
ENSDART00000124724 |
dpydb
|
dihydropyrimidine dehydrogenase b |
| chr7_-_51953807 | 0.34 |
ENSDART00000174102
ENSDART00000145645 ENSDART00000052054 |
cyp2x10.2
|
cytochrome P450, family 2, subfamily X, polypeptide 10.2 |
| chr2_+_3533117 | 0.34 |
ENSDART00000132572
|
gpt2l
|
glutamic pyruvate transaminase (alanine aminotransferase) 2, like |
| chr9_-_33121535 | 0.34 |
ENSDART00000166371
ENSDART00000138052 |
zgc:172014
|
zgc:172014 |
| chr4_-_63558838 | 0.34 |
ENSDART00000191708
ENSDART00000190740 ENSDART00000186476 |
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr3_+_12710350 | 0.34 |
ENSDART00000157959
|
cyp2k18
|
cytochrome P450, family 2, subfamily K, polypeptide 18 |
| chr4_+_34119824 | 0.33 |
ENSDART00000165637
|
si:ch211-223g7.6
|
si:ch211-223g7.6 |
| chr18_-_37007061 | 0.33 |
ENSDART00000136432
|
map3k10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr6_-_1587291 | 0.33 |
ENSDART00000067592
ENSDART00000178877 |
zgc:123305
|
zgc:123305 |
| chr18_-_158541 | 0.33 |
ENSDART00000188914
ENSDART00000191052 |
trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr12_-_14551077 | 0.33 |
ENSDART00000188717
|
FO704673.1
|
|
| chr9_+_24936496 | 0.33 |
ENSDART00000157474
|
slc39a10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr4_-_36139585 | 0.33 |
ENSDART00000132071
|
znf992
|
zinc finger protein 992 |
| chr23_-_46040618 | 0.32 |
ENSDART00000161415
|
CABZ01080918.1
|
|
| chr21_+_1119046 | 0.31 |
ENSDART00000184678
|
CABZ01088049.1
|
|
| chr22_-_20814450 | 0.31 |
ENSDART00000089076
|
dot1l
|
DOT1-like histone H3K79 methyltransferase |
| chr11_+_6115621 | 0.30 |
ENSDART00000165031
ENSDART00000027666 ENSDART00000161458 |
nr2f6b
|
nuclear receptor subfamily 2, group F, member 6b |
| chr18_+_45200043 | 0.30 |
ENSDART00000015786
|
large2
|
LARGE xylosyl- and glucuronyltransferase 2 |
| chr18_-_39200557 | 0.30 |
ENSDART00000132367
ENSDART00000183672 |
si:ch211-235f12.2
mapk6
|
si:ch211-235f12.2 mitogen-activated protein kinase 6 |
| chr1_-_52292235 | 0.30 |
ENSDART00000132638
|
si:dkey-121b10.7
|
si:dkey-121b10.7 |
| chr21_+_7188943 | 0.30 |
ENSDART00000172174
|
agpat2
|
1-acylglycerol-3-phosphate O-acyltransferase 2 (lysophosphatidic acid acyltransferase, beta) |
| chr17_-_6076084 | 0.30 |
ENSDART00000058890
|
ephx2
|
epoxide hydrolase 2, cytoplasmic |
| chr12_-_6033824 | 0.30 |
ENSDART00000131301
ENSDART00000139419 ENSDART00000032050 |
g6pca.1
|
glucose-6-phosphatase a, catalytic subunit, tandem duplicate 1 |
| chr5_-_62317496 | 0.30 |
ENSDART00000180089
|
zgc:85789
|
zgc:85789 |
| chr13_-_33114933 | 0.29 |
ENSDART00000140543
ENSDART00000075953 |
zfyve26
|
zinc finger, FYVE domain containing 26 |
| chr12_+_30368145 | 0.29 |
ENSDART00000153454
|
ccdc186
|
si:ch211-225b10.4 |
| chr7_-_52096498 | 0.29 |
ENSDART00000098688
ENSDART00000098690 |
cyp2x10.2
|
cytochrome P450, family 2, subfamily X, polypeptide 10.2 |
| chr9_-_22822084 | 0.29 |
ENSDART00000142020
|
neb
|
nebulin |
| chr23_-_24493768 | 0.29 |
ENSDART00000135559
ENSDART00000046933 |
sult1st5
|
sulfotransferase family 1, cytosolic sulfotransferase 5 |
| chr4_+_53246788 | 0.29 |
ENSDART00000184708
ENSDART00000169256 |
si:dkey-250k10.1
|
si:dkey-250k10.1 |
| chr5_-_24238733 | 0.29 |
ENSDART00000138170
|
plscr3a
|
phospholipid scramblase 3a |
| chr25_+_34247107 | 0.28 |
ENSDART00000148507
|
bnip2
|
BCL2 interacting protein 2 |
| chr9_+_54984537 | 0.28 |
ENSDART00000029528
|
mospd2
|
motile sperm domain containing 2 |
| chr16_+_11558868 | 0.28 |
ENSDART00000112497
ENSDART00000180445 |
zgc:198329
|
zgc:198329 |
| chr21_+_40944530 | 0.28 |
ENSDART00000022976
|
kctd16b
|
potassium channel tetramerization domain containing 16b |
| chr4_+_69619 | 0.28 |
ENSDART00000164425
|
mansc1
|
MANSC domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr2f2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:0006549 | isoleucine metabolic process(GO:0006549) isoleucine biosynthetic process(GO:0009097) |
| 0.4 | 1.1 | GO:2000193 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.3 | 1.3 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.3 | 0.9 | GO:0009750 | response to fructose(GO:0009750) |
| 0.2 | 0.7 | GO:0072679 | thymocyte migration(GO:0072679) |
| 0.2 | 0.9 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.7 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.1 | 0.7 | GO:0002164 | larval development(GO:0002164) larval heart development(GO:0007508) angiogenesis involved in wound healing(GO:0060055) |
| 0.1 | 0.5 | GO:0071871 | response to monoamine(GO:0071867) response to catecholamine(GO:0071869) response to epinephrine(GO:0071871) |
| 0.1 | 0.4 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.1 | 0.5 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 0.1 | 0.6 | GO:0042264 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.6 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 0.3 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.1 | 0.3 | GO:0046125 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.1 | 0.5 | GO:0032530 | regulation of microvillus organization(GO:0032530) regulation of microvillus assembly(GO:0032534) |
| 0.1 | 1.0 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 0.3 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 | 0.7 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.1 | 1.2 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.1 | 0.5 | GO:0052651 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.1 | 0.3 | GO:0005991 | trehalose metabolic process(GO:0005991) |
| 0.1 | 0.3 | GO:0046689 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 0.1 | 1.4 | GO:1901642 | nucleoside transport(GO:0015858) nucleoside transmembrane transport(GO:1901642) |
| 0.1 | 4.7 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.1 | 0.3 | GO:1990575 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.1 | 0.3 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.1 | 0.3 | GO:0034729 | histone H3-K79 methylation(GO:0034729) regulation of transcription regulatory region DNA binding(GO:2000677) |
| 0.1 | 0.8 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.1 | 0.4 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.1 | 0.2 | GO:0019483 | beta-alanine metabolic process(GO:0019482) beta-alanine biosynthetic process(GO:0019483) |
| 0.1 | 0.4 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.1 | 0.4 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 2.6 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.1 | 0.7 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.5 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 0.4 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.1 | 0.4 | GO:0097107 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.1 | 0.2 | GO:0072020 | proximal straight tubule development(GO:0072020) |
| 0.0 | 0.8 | GO:1903286 | positive regulation of sodium ion transport(GO:0010765) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 1.0 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.3 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 1.2 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 1.3 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.3 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.0 | 0.1 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.0 | 0.6 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 1.5 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.5 | GO:0050482 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.1 | GO:0071548 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) short-chain fatty acid catabolic process(GO:0019626) response to dexamethasone(GO:0071548) |
| 0.0 | 0.5 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.0 | 1.2 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 0.7 | GO:0007257 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.8 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 0.6 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.5 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.6 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.8 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.7 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.4 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.2 | GO:0016576 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.2 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.2 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.1 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.1 | GO:0061178 | insulin secretion involved in cellular response to glucose stimulus(GO:0035773) regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.0 | 0.6 | GO:0060841 | venous blood vessel development(GO:0060841) |
| 0.0 | 0.2 | GO:0016139 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.0 | 0.1 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.0 | 0.3 | GO:0042396 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 0.1 | GO:0009078 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.0 | 0.1 | GO:0098920 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.0 | 0.4 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.3 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.3 | GO:0046473 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.1 | GO:1905048 | regulation of metallopeptidase activity(GO:1905048) |
| 0.0 | 0.8 | GO:0031647 | regulation of protein stability(GO:0031647) |
| 0.0 | 1.2 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.2 | GO:0042493 | response to drug(GO:0042493) |
| 0.0 | 0.6 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.2 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.1 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.1 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.3 | GO:0048679 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 0.1 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.9 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.6 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.1 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.0 | 0.8 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.0 | 0.7 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.2 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.2 | 0.7 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.2 | 0.6 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 0.4 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 0.5 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 0.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.7 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.5 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.6 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 1.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.4 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.7 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.6 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.6 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.7 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.2 | 0.6 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.2 | 0.6 | GO:0015228 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.2 | 0.7 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.2 | 2.0 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.2 | 2.4 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.2 | 0.6 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.1 | 0.7 | GO:0051430 | mu-type opioid receptor binding(GO:0031852) dopamine receptor binding(GO:0050780) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 1.0 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.5 | GO:0015105 | arsenite transmembrane transporter activity(GO:0015105) |
| 0.1 | 0.9 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 1.2 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 1.2 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.1 | 1.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 1.1 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 0.3 | GO:0015927 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 0.1 | 0.4 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.1 | 0.8 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 5.5 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 0.3 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.1 | 0.7 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.3 | GO:0031151 | histone methyltransferase activity (H3-K79 specific)(GO:0031151) |
| 0.1 | 0.4 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 0.3 | GO:0002058 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.1 | 0.6 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 0.6 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 0.5 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.1 | 0.6 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 0.7 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 0.5 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.2 | GO:0004557 | alpha-galactosidase activity(GO:0004557) |
| 0.1 | 0.4 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 1.5 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.5 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.0 | 0.8 | GO:0019870 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.6 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 2.6 | GO:0015370 | solute:sodium symporter activity(GO:0015370) |
| 0.0 | 0.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 2.1 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.5 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 2.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.3 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.6 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.3 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.2 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.3 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.3 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.3 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.5 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.9 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.5 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.6 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.3 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.1 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.0 | 0.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.2 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 1.1 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.1 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.1 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.1 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.6 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.3 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.5 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.5 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.4 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.6 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.7 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.6 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.7 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.4 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.2 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.5 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.8 | PID P73PATHWAY | p73 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 0.7 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.1 | 1.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.2 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.6 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.0 | 0.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.4 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.6 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.1 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |