Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
Results for nr2f1a
Z-value: 0.92
Transcription factors associated with nr2f1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr2f1a
|
ENSDARG00000052695 | nuclear receptor subfamily 2, group F, member 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr2f1a | dr11_v1_chr5_+_49744713_49744713 | -0.80 | 4.2e-05 | Click! |
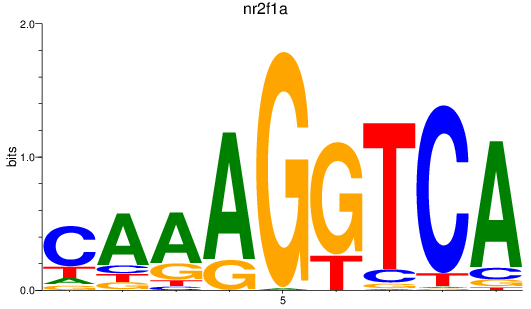
Activity profile of nr2f1a motif
Sorted Z-values of nr2f1a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_35068036 | 4.23 |
ENSDART00000022139
|
zgc:136461
|
zgc:136461 |
| chr13_-_12581388 | 2.30 |
ENSDART00000079655
|
enpep
|
glutamyl aminopeptidase |
| chr21_+_25226558 | 2.24 |
ENSDART00000168480
|
sycn.2
|
syncollin, tandem duplicate 2 |
| chr16_+_26012569 | 2.22 |
ENSDART00000148846
|
prss59.1
|
protease, serine, 59, tandem duplicate 1 |
| chr1_-_50838160 | 1.90 |
ENSDART00000163939
ENSDART00000165111 |
zgc:154142
|
zgc:154142 |
| chr10_-_11385155 | 1.85 |
ENSDART00000064214
|
plac8.1
|
placenta-specific 8, tandem duplicate 1 |
| chr3_+_7771420 | 1.84 |
ENSDART00000156809
ENSDART00000156309 |
hook2
|
hook microtubule-tethering protein 2 |
| chr23_+_42434348 | 1.65 |
ENSDART00000161027
|
cyp2aa1
|
cytochrome P450, family 2, subfamily AA, polypeptide 1 |
| chr7_-_24005268 | 1.62 |
ENSDART00000173608
|
si:dkey-183c6.9
|
si:dkey-183c6.9 |
| chr11_-_8167799 | 1.57 |
ENSDART00000133574
ENSDART00000024046 ENSDART00000146940 |
uox
|
urate oxidase |
| chr21_+_25216397 | 1.55 |
ENSDART00000130970
|
sycn.3
|
syncollin, tandem duplicate 3 |
| chr16_-_2558653 | 1.44 |
ENSDART00000110365
|
adcy3a
|
adenylate cyclase 3a |
| chr17_-_15149192 | 1.43 |
ENSDART00000180511
ENSDART00000103405 |
gch1
|
GTP cyclohydrolase 1 |
| chr19_-_703898 | 1.42 |
ENSDART00000181096
ENSDART00000121462 |
slc6a19a.2
|
solute carrier family 6 (neutral amino acid transporter), member 19a, tandem duplicate 2 |
| chr13_-_303137 | 1.39 |
ENSDART00000099131
|
chs1
|
chitin synthase 1 |
| chr10_-_32524771 | 1.37 |
ENSDART00000066793
|
dgat2
|
diacylglycerol O-acyltransferase 2 |
| chr22_+_38778649 | 1.34 |
ENSDART00000075873
|
alpi.2
|
alkaline phosphatase, intestinal, tandem duplicate 2 |
| chr6_-_345503 | 1.32 |
ENSDART00000168901
|
pde6ha
|
phosphodiesterase 6H, cGMP-specific, cone, gamma, paralog a |
| chr25_-_30429607 | 1.32 |
ENSDART00000162429
ENSDART00000176535 |
si:ch211-93f2.1
|
si:ch211-93f2.1 |
| chr7_-_20758825 | 1.31 |
ENSDART00000156717
ENSDART00000182629 ENSDART00000179801 |
chd3
|
chromodomain helicase DNA binding protein 3 |
| chr5_-_36549024 | 1.27 |
ENSDART00000097671
|
zgc:158432
|
zgc:158432 |
| chr6_-_49873020 | 1.26 |
ENSDART00000148511
|
gnas
|
GNAS complex locus |
| chr10_+_45128375 | 1.24 |
ENSDART00000164805
|
camk2b2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 2 |
| chr7_-_26457208 | 1.24 |
ENSDART00000173519
|
zgc:172079
|
zgc:172079 |
| chr17_-_25737452 | 1.21 |
ENSDART00000152021
|
si:ch211-214p16.3
|
si:ch211-214p16.3 |
| chr25_-_25384045 | 1.21 |
ENSDART00000150631
|
zgc:123278
|
zgc:123278 |
| chr22_+_38778854 | 1.20 |
ENSDART00000182926
ENSDART00000125466 |
alpi.2
|
alkaline phosphatase, intestinal, tandem duplicate 2 |
| chr2_-_53592532 | 1.16 |
ENSDART00000184066
|
ccl25a
|
chemokine (C-C motif) ligand 25a |
| chr24_-_29586082 | 1.12 |
ENSDART00000136763
|
vav3a
|
vav 3 guanine nucleotide exchange factor a |
| chr1_-_9249943 | 1.11 |
ENSDART00000055011
|
zgc:136472
|
zgc:136472 |
| chr5_+_1278092 | 1.06 |
ENSDART00000147972
ENSDART00000159783 |
dnm1a
|
dynamin 1a |
| chr6_-_39344259 | 1.05 |
ENSDART00000104074
|
zgc:158846
|
zgc:158846 |
| chr12_-_31457301 | 1.04 |
ENSDART00000043887
ENSDART00000148603 |
acsl5
|
acyl-CoA synthetase long chain family member 5 |
| chr2_+_52232630 | 1.03 |
ENSDART00000006216
|
plpp2a
|
phospholipid phosphatase 2a |
| chr8_+_43053519 | 1.03 |
ENSDART00000147178
|
prnpa
|
prion protein a |
| chr8_-_50482781 | 1.00 |
ENSDART00000056361
|
ido1
|
indoleamine 2,3-dioxygenase 1 |
| chr9_-_48736388 | 0.99 |
ENSDART00000022074
|
dhrs9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr5_+_8919698 | 0.98 |
ENSDART00000046440
|
agpat9l
|
1-acylglycerol-3-phosphate O-acyltransferase 9, like |
| chr8_-_12468744 | 0.97 |
ENSDART00000135019
|
FIBCD1 (1 of many)
|
si:dkeyp-51b7.3 |
| chr17_+_12408405 | 0.97 |
ENSDART00000154827
ENSDART00000048440 ENSDART00000156429 |
khk
|
ketohexokinase |
| chr12_-_20362041 | 0.97 |
ENSDART00000184145
ENSDART00000105952 |
aqp8a.2
|
aquaporin 8a, tandem duplicate 2 |
| chr12_+_22607761 | 0.94 |
ENSDART00000153112
|
si:dkey-219e21.2
|
si:dkey-219e21.2 |
| chr11_+_21910343 | 0.92 |
ENSDART00000161485
|
foxp4
|
forkhead box P4 |
| chr11_+_36989696 | 0.91 |
ENSDART00000045888
|
tkta
|
transketolase a |
| chr25_-_31898552 | 0.91 |
ENSDART00000156128
|
si:ch73-330k17.3
|
si:ch73-330k17.3 |
| chr24_-_11127493 | 0.90 |
ENSDART00000144066
|
myrip
|
myosin VIIA and Rab interacting protein |
| chr19_-_20093341 | 0.90 |
ENSDART00000129917
|
mpp6b
|
membrane protein, palmitoylated 6b (MAGUK p55 subfamily member 6) |
| chr20_+_43925266 | 0.89 |
ENSDART00000037379
|
clic5b
|
chloride intracellular channel 5b |
| chr7_+_14005111 | 0.89 |
ENSDART00000187365
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr8_-_33154677 | 0.89 |
ENSDART00000133300
|
zbtb34
|
zinc finger and BTB domain containing 34 |
| chr3_-_58455289 | 0.88 |
ENSDART00000052179
|
cdr2a
|
cerebellar degeneration-related protein 2a |
| chr5_-_23280098 | 0.88 |
ENSDART00000126540
ENSDART00000051533 |
plp1b
|
proteolipid protein 1b |
| chr22_+_19290199 | 0.87 |
ENSDART00000148173
|
si:dkey-21e2.15
|
si:dkey-21e2.15 |
| chr9_+_9502610 | 0.87 |
ENSDART00000061525
ENSDART00000125174 |
nr1i2
|
nuclear receptor subfamily 1, group I, member 2 |
| chr20_-_1141722 | 0.86 |
ENSDART00000152675
|
gabrr1
|
gamma-aminobutyric acid (GABA) A receptor, rho 1 |
| chr23_+_21544227 | 0.86 |
ENSDART00000140253
|
arhgef10lb
|
Rho guanine nucleotide exchange factor (GEF) 10-like b |
| chr11_+_5681762 | 0.86 |
ENSDART00000179139
|
arid3a
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr3_+_30257582 | 0.85 |
ENSDART00000159497
ENSDART00000103457 ENSDART00000121883 |
mybpc2a
|
myosin binding protein C, fast type a |
| chr20_+_25575391 | 0.85 |
ENSDART00000063108
|
cyp2p8
|
cytochrome P450, family 2, subfamily P, polypeptide 8 |
| chr12_-_6172154 | 0.84 |
ENSDART00000185434
|
a1cf
|
apobec1 complementation factor |
| chr9_-_44905867 | 0.83 |
ENSDART00000138316
ENSDART00000131252 ENSDART00000179383 ENSDART00000159337 |
zgc:66484
|
zgc:66484 |
| chr23_-_45504991 | 0.83 |
ENSDART00000148761
|
col24a1
|
collagen type XXIV alpha 1 |
| chr16_-_34258931 | 0.83 |
ENSDART00000145485
|
phactr4b
|
phosphatase and actin regulator 4b |
| chr18_+_45542981 | 0.83 |
ENSDART00000140357
|
kifc3
|
kinesin family member C3 |
| chr18_+_44768829 | 0.83 |
ENSDART00000016271
|
ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr21_+_25221940 | 0.82 |
ENSDART00000108972
|
sycn.1
|
syncollin, tandem duplicate 1 |
| chr7_+_73649686 | 0.82 |
ENSDART00000185589
|
PCP4L1
|
si:dkey-46i9.1 |
| chr1_-_46343999 | 0.81 |
ENSDART00000145117
ENSDART00000193233 |
atp11a
|
ATPase phospholipid transporting 11A |
| chr23_+_40461411 | 0.81 |
ENSDART00000184223
|
soga3b
|
SOGA family member 3b |
| chr15_-_33904831 | 0.81 |
ENSDART00000164333
ENSDART00000165404 |
mag
|
myelin associated glycoprotein |
| chr13_-_31544365 | 0.80 |
ENSDART00000005670
|
dhrs7
|
dehydrogenase/reductase (SDR family) member 7 |
| chr9_-_42861080 | 0.80 |
ENSDART00000193688
|
ttn.1
|
titin, tandem duplicate 1 |
| chr9_-_40846447 | 0.79 |
ENSDART00000143384
|
si:dkey-95p16.1
|
si:dkey-95p16.1 |
| chr23_+_27912079 | 0.79 |
ENSDART00000171859
|
BX539336.1
|
|
| chr2_+_37110504 | 0.78 |
ENSDART00000132794
ENSDART00000042974 |
slc1a8b
|
solute carrier family 1 (glutamate transporter), member 8b |
| chr21_-_43550120 | 0.78 |
ENSDART00000151627
|
si:ch73-362m14.2
|
si:ch73-362m14.2 |
| chr20_+_18163355 | 0.77 |
ENSDART00000011287
|
aqp4
|
aquaporin 4 |
| chr18_+_44769027 | 0.76 |
ENSDART00000145190
|
ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr8_-_53108207 | 0.76 |
ENSDART00000111023
|
b3galt4
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 4 |
| chr25_+_35706493 | 0.75 |
ENSDART00000176741
|
kif21a
|
kinesin family member 21A |
| chr7_-_24520866 | 0.75 |
ENSDART00000077039
|
faah2b
|
fatty acid amide hydrolase 2b |
| chr9_-_30346279 | 0.75 |
ENSDART00000089488
|
sytl5
|
synaptotagmin-like 5 |
| chr10_-_35108683 | 0.74 |
ENSDART00000049633
|
zgc:110006
|
zgc:110006 |
| chr17_+_12408188 | 0.73 |
ENSDART00000105218
|
khk
|
ketohexokinase |
| chr12_-_48943467 | 0.73 |
ENSDART00000191829
|
CABZ01092907.1
|
|
| chr5_-_20678300 | 0.73 |
ENSDART00000088639
|
wscd2
|
WSC domain containing 2 |
| chr11_-_34147205 | 0.73 |
ENSDART00000173216
|
atp13a3
|
ATPase 13A3 |
| chr6_-_49547680 | 0.73 |
ENSDART00000169678
|
ppp4r1l
|
protein phosphatase 4, regulatory subunit 1-like |
| chr24_-_14711597 | 0.72 |
ENSDART00000131830
|
jph1a
|
junctophilin 1a |
| chr5_-_69934558 | 0.71 |
ENSDART00000124954
|
ugt2a4
|
UDP glucuronosyltransferase 2 family, polypeptide A4 |
| chr2_+_43755274 | 0.71 |
ENSDART00000138620
|
arhgap12a
|
Rho GTPase activating protein 12a |
| chr3_-_1317290 | 0.70 |
ENSDART00000047094
|
LO018552.1
|
|
| chr16_+_53603945 | 0.70 |
ENSDART00000083513
|
sacm1lb
|
SAC1 like phosphatidylinositide phosphatase b |
| chr10_-_41157135 | 0.70 |
ENSDART00000134851
|
aak1b
|
AP2 associated kinase 1b |
| chr1_+_25848231 | 0.70 |
ENSDART00000027973
|
fabp2
|
fatty acid binding protein 2, intestinal |
| chr25_-_35599887 | 0.69 |
ENSDART00000153827
|
clpxb
|
caseinolytic mitochondrial matrix peptidase chaperone subunit b |
| chr13_-_11644806 | 0.69 |
ENSDART00000169953
|
dctn1b
|
dynactin 1b |
| chr24_-_34034931 | 0.68 |
ENSDART00000169227
|
asic1c
|
acid-sensing (proton-gated) ion channel 1c |
| chr1_+_44826367 | 0.68 |
ENSDART00000146962
|
STX3
|
zgc:165520 |
| chr6_+_30703828 | 0.68 |
ENSDART00000104628
|
insl5a
|
insulin-like 5a |
| chr15_-_16884912 | 0.67 |
ENSDART00000062135
|
zgc:103681
|
zgc:103681 |
| chr2_+_7192966 | 0.67 |
ENSDART00000142735
|
si:ch211-13f8.1
|
si:ch211-13f8.1 |
| chr19_-_42045372 | 0.67 |
ENSDART00000144275
|
trioa
|
trio Rho guanine nucleotide exchange factor a |
| chr24_-_39518599 | 0.67 |
ENSDART00000145606
ENSDART00000031486 |
lyrm1
|
LYR motif containing 1 |
| chr3_+_22905341 | 0.67 |
ENSDART00000111435
|
hdac5
|
histone deacetylase 5 |
| chr25_+_14092871 | 0.67 |
ENSDART00000067239
|
guca1g
|
guanylate cyclase activator 1g |
| chr25_-_13614702 | 0.67 |
ENSDART00000165510
ENSDART00000190959 |
fa2h
|
fatty acid 2-hydroxylase |
| chr25_-_35107791 | 0.66 |
ENSDART00000192819
|
zgc:165555
|
zgc:165555 |
| chr7_+_73332486 | 0.66 |
ENSDART00000174119
ENSDART00000092051 ENSDART00000192388 |
CABZ01081780.1
|
|
| chr22_+_7486867 | 0.66 |
ENSDART00000034586
|
CELA1 (1 of many)
|
zgc:112302 |
| chr23_-_42810664 | 0.66 |
ENSDART00000102328
|
pfkfb2a
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2a |
| chr20_+_51006362 | 0.66 |
ENSDART00000028084
|
mpp5b
|
membrane protein, palmitoylated 5b (MAGUK p55 subfamily member 5) |
| chr18_+_5273953 | 0.65 |
ENSDART00000165073
|
slc12a1
|
solute carrier family 12 (sodium/potassium/chloride transporter), member 1 |
| chr23_+_42810055 | 0.65 |
ENSDART00000186647
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr18_+_27926839 | 0.65 |
ENSDART00000191835
|
hipk3b
|
homeodomain interacting protein kinase 3b |
| chr21_+_5589923 | 0.65 |
ENSDART00000160885
|
stbd1
|
starch binding domain 1 |
| chr1_+_58094551 | 0.65 |
ENSDART00000146316
|
si:ch211-114l13.1
|
si:ch211-114l13.1 |
| chr15_+_20403903 | 0.64 |
ENSDART00000134182
|
cox7a1
|
cytochrome c oxidase subunit VIIa polypeptide 1 (muscle) |
| chr23_-_27701361 | 0.64 |
ENSDART00000186688
ENSDART00000183985 |
dnajc22
|
DnaJ (Hsp40) homolog, subfamily C, member 22 |
| chr17_-_30702411 | 0.64 |
ENSDART00000114358
|
zgc:194392
|
zgc:194392 |
| chr11_+_33284837 | 0.64 |
ENSDART00000166239
ENSDART00000111412 |
nkpd1
|
NTPase, KAP family P-loop domain containing 1 |
| chr5_-_57879138 | 0.64 |
ENSDART00000145959
|
sik2a
|
salt-inducible kinase 2a |
| chr24_-_2843107 | 0.63 |
ENSDART00000165290
|
cyb5a
|
cytochrome b5 type A (microsomal) |
| chr25_-_26018240 | 0.63 |
ENSDART00000150800
|
acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr22_-_16403602 | 0.63 |
ENSDART00000183417
|
lama3
|
laminin, alpha 3 |
| chr2_-_57076687 | 0.63 |
ENSDART00000161523
|
slc25a42
|
solute carrier family 25, member 42 |
| chr25_-_19420949 | 0.62 |
ENSDART00000181338
|
map1ab
|
microtubule-associated protein 1Ab |
| chr6_+_42587637 | 0.62 |
ENSDART00000179964
|
camkva
|
CaM kinase-like vesicle-associated a |
| chr3_+_12744083 | 0.62 |
ENSDART00000158554
ENSDART00000169545 |
cyp2k21
|
cytochrome P450, family 2, subfamily k, polypeptide 21 |
| chr13_-_37254777 | 0.62 |
ENSDART00000139734
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr12_+_48972915 | 0.62 |
ENSDART00000170695
|
lrit1b
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 1b |
| chr24_-_24271629 | 0.61 |
ENSDART00000135060
|
rps6ka3b
|
ribosomal protein S6 kinase, polypeptide 3b |
| chr15_-_21239416 | 0.61 |
ENSDART00000155787
|
A2ML1 (1 of many)
|
si:dkey-105h12.2 |
| chr10_+_39164638 | 0.61 |
ENSDART00000188997
|
CU633908.1
|
|
| chr15_+_29393519 | 0.60 |
ENSDART00000193488
ENSDART00000112375 |
gdpd5b
|
glycerophosphodiester phosphodiesterase domain containing 5b |
| chr8_+_6576940 | 0.60 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr17_-_47142249 | 0.60 |
ENSDART00000184705
|
CU915775.1
|
|
| chr15_-_1843831 | 0.60 |
ENSDART00000156718
ENSDART00000154175 |
taf15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr5_+_57328535 | 0.59 |
ENSDART00000149646
|
slc31a1
|
solute carrier family 31 (copper transporter), member 1 |
| chr6_-_53204808 | 0.59 |
ENSDART00000160955
|
slc38a3b
|
solute carrier family 38, member 3b |
| chr24_-_26369185 | 0.59 |
ENSDART00000080039
|
lrrc31
|
leucine rich repeat containing 31 |
| chr8_-_23780334 | 0.59 |
ENSDART00000145179
ENSDART00000145894 |
zgc:195245
|
zgc:195245 |
| chr18_+_50461981 | 0.58 |
ENSDART00000158761
|
CU896640.1
|
|
| chr6_-_48087152 | 0.58 |
ENSDART00000180614
|
slc2a1b
|
solute carrier family 2 (facilitated glucose transporter), member 1b |
| chr18_+_7309150 | 0.57 |
ENSDART00000052792
|
cers3b
|
ceramide synthase 3b |
| chr2_-_8102169 | 0.57 |
ENSDART00000131955
|
pls1
|
plastin 1 (I isoform) |
| chr15_+_45640906 | 0.57 |
ENSDART00000149361
ENSDART00000149079 |
sagb
|
S-antigen; retina and pineal gland (arrestin) b |
| chr3_-_37351225 | 0.57 |
ENSDART00000174685
|
si:ch211-278a6.1
|
si:ch211-278a6.1 |
| chr8_-_45834825 | 0.57 |
ENSDART00000132965
|
ogdha
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase a (lipoamide) |
| chr8_-_45835056 | 0.56 |
ENSDART00000022242
|
ogdha
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase a (lipoamide) |
| chr17_-_29194219 | 0.56 |
ENSDART00000157340
|
sptbn5
|
spectrin, beta, non-erythrocytic 5 |
| chr3_+_52737565 | 0.56 |
ENSDART00000108639
|
gmip
|
GEM interacting protein |
| chr22_-_17953529 | 0.56 |
ENSDART00000135716
|
ncanb
|
neurocan b |
| chr6_+_55006686 | 0.55 |
ENSDART00000081703
|
nav1b
|
neuron navigator 1b |
| chr7_-_20731078 | 0.55 |
ENSDART00000188267
|
chd3
|
chromodomain helicase DNA binding protein 3 |
| chr20_+_43379029 | 0.55 |
ENSDART00000142486
ENSDART00000186486 |
unc93a
|
unc-93 homolog A |
| chr15_+_28119284 | 0.55 |
ENSDART00000188366
|
unc119a
|
unc-119 homolog a (C. elegans) |
| chr18_-_16791331 | 0.55 |
ENSDART00000148222
|
ampd3b
|
adenosine monophosphate deaminase 3b |
| chr16_+_25407021 | 0.54 |
ENSDART00000187489
ENSDART00000086333 |
jarid2a
|
jumonji, AT rich interactive domain 2a |
| chr11_-_45141309 | 0.54 |
ENSDART00000181736
|
cant1b
|
calcium activated nucleotidase 1b |
| chr16_+_14033121 | 0.54 |
ENSDART00000135844
|
rusc1
|
RUN and SH3 domain containing 1 |
| chr16_-_13109749 | 0.54 |
ENSDART00000142610
|
prkcg
|
protein kinase C, gamma |
| chr6_-_10320676 | 0.54 |
ENSDART00000151247
|
scn1lab
|
sodium channel, voltage-gated, type I like, alpha b |
| chr8_+_36339470 | 0.53 |
ENSDART00000191250
|
FO744840.1
|
|
| chr1_-_19215336 | 0.53 |
ENSDART00000162949
ENSDART00000170680 |
ptprdb
|
protein tyrosine phosphatase, receptor type, D, b |
| chr3_+_56876280 | 0.52 |
ENSDART00000154197
|
amn
|
amnion associated transmembrane protein |
| chr4_+_15944245 | 0.52 |
ENSDART00000134594
|
si:dkey-117n7.3
|
si:dkey-117n7.3 |
| chr16_-_17175731 | 0.52 |
ENSDART00000183057
|
opn9
|
opsin 9 |
| chr2_-_8111346 | 0.51 |
ENSDART00000123103
|
pls1
|
plastin 1 (I isoform) |
| chr5_-_68927728 | 0.51 |
ENSDART00000132838
|
ank1a
|
ankyrin 1, erythrocytic a |
| chr21_-_21096437 | 0.51 |
ENSDART00000186552
|
ank1b
|
ankyrin 1, erythrocytic b |
| chr8_+_8726958 | 0.51 |
ENSDART00000110854
|
elk1
|
ELK1, member of ETS oncogene family |
| chr15_+_29292154 | 0.51 |
ENSDART00000137817
|
rap1gap2a
|
RAP1 GTPase activating protein 2a |
| chr14_+_33413980 | 0.50 |
ENSDART00000052780
ENSDART00000124437 ENSDART00000173327 |
ndufa1
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 1 |
| chr3_+_12710350 | 0.50 |
ENSDART00000157959
|
cyp2k18
|
cytochrome P450, family 2, subfamily K, polypeptide 18 |
| chr7_+_13988075 | 0.50 |
ENSDART00000186812
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr5_+_65493699 | 0.50 |
ENSDART00000161567
|
si:dkey-21e5.1
|
si:dkey-21e5.1 |
| chr16_+_26774182 | 0.50 |
ENSDART00000042895
|
cdh17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr4_+_25917915 | 0.49 |
ENSDART00000138603
|
vezt
|
vezatin, adherens junctions transmembrane protein |
| chr2_-_23778180 | 0.49 |
ENSDART00000136782
|
si:dkey-24c2.7
|
si:dkey-24c2.7 |
| chr23_+_4583747 | 0.49 |
ENSDART00000160350
|
LO017700.1
|
|
| chr6_+_60055168 | 0.49 |
ENSDART00000008752
|
pck1
|
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr12_-_47601845 | 0.49 |
ENSDART00000169548
ENSDART00000182889 |
rgs7b
|
regulator of G protein signaling 7b |
| chr20_-_51917771 | 0.49 |
ENSDART00000147240
|
dusp10
|
dual specificity phosphatase 10 |
| chr7_+_27251376 | 0.49 |
ENSDART00000173521
ENSDART00000173962 |
sox6
|
SRY (sex determining region Y)-box 6 |
| chr13_-_21739142 | 0.48 |
ENSDART00000078460
|
si:dkey-191g9.5
|
si:dkey-191g9.5 |
| chr21_+_40944530 | 0.48 |
ENSDART00000022976
|
kctd16b
|
potassium channel tetramerization domain containing 16b |
| chr5_-_13076779 | 0.48 |
ENSDART00000192826
|
ypel1
|
yippee-like 1 |
| chr19_-_3303995 | 0.48 |
ENSDART00000105150
|
si:ch211-133n4.9
|
si:ch211-133n4.9 |
| chr11_-_22997506 | 0.48 |
ENSDART00000167817
|
atp2b2
|
ATPase plasma membrane Ca2+ transporting 2 |
| chr21_-_1972236 | 0.48 |
ENSDART00000192858
ENSDART00000189962 ENSDART00000182461 ENSDART00000165547 |
WDR7
|
WD repeat domain 7 |
| chr3_-_18373425 | 0.47 |
ENSDART00000178522
|
spag9a
|
sperm associated antigen 9a |
| chr18_-_12957451 | 0.47 |
ENSDART00000140403
|
srgap1a
|
SLIT-ROBO Rho GTPase activating protein 1a |
| chr15_+_47386939 | 0.47 |
ENSDART00000128224
|
FO904873.1
|
|
| chr2_+_10127762 | 0.47 |
ENSDART00000100726
|
insl5b
|
insulin-like 5b |
| chr16_-_47483142 | 0.47 |
ENSDART00000147072
|
cthrc1b
|
collagen triple helix repeat containing 1b |
| chr4_+_43700319 | 0.47 |
ENSDART00000141967
|
si:ch211-226o13.1
|
si:ch211-226o13.1 |
| chr8_+_8012570 | 0.47 |
ENSDART00000183429
|
si:ch211-169p10.1
|
si:ch211-169p10.1 |
| chr23_-_637347 | 0.47 |
ENSDART00000132175
|
l1camb
|
L1 cell adhesion molecule, paralog b |
| chr18_+_9382847 | 0.46 |
ENSDART00000061886
|
sema3ab
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Ab |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr2f1a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0009750 | response to fructose(GO:0009750) |
| 0.4 | 1.6 | GO:0006549 | isoleucine metabolic process(GO:0006549) isoleucine biosynthetic process(GO:0009097) |
| 0.4 | 1.2 | GO:0072679 | thymocyte migration(GO:0072679) |
| 0.4 | 1.4 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.3 | 1.0 | GO:2000193 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.3 | 1.4 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.3 | 1.4 | GO:1901073 | chitin biosynthetic process(GO:0006031) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.3 | 1.3 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.2 | 1.3 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.2 | 0.9 | GO:0070316 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.2 | 0.6 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.2 | 0.6 | GO:0019628 | urate catabolic process(GO:0019628) urate metabolic process(GO:0046415) |
| 0.2 | 0.9 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.2 | 0.5 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 0.6 | GO:0006867 | asparagine transport(GO:0006867) |
| 0.1 | 0.9 | GO:0038065 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.1 | 0.6 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.1 | 0.5 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.1 | 0.4 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.1 | 1.0 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.1 | 0.5 | GO:0019401 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) short-chain fatty acid catabolic process(GO:0019626) response to dexamethasone(GO:0071548) |
| 0.1 | 0.7 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.1 | 0.6 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.1 | 0.8 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 1.0 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 1.4 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.1 | 0.4 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.1 | 0.7 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 0.5 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 0.4 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.1 | 0.3 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.1 | 0.8 | GO:0006833 | water transport(GO:0006833) |
| 0.1 | 0.3 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.1 | 0.4 | GO:0019482 | beta-alanine metabolic process(GO:0019482) beta-alanine biosynthetic process(GO:0019483) |
| 0.1 | 2.3 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.1 | 0.2 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.1 | 0.2 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.1 | 0.2 | GO:0015882 | L-ascorbic acid transport(GO:0015882) |
| 0.1 | 0.5 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.1 | 1.0 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 0.3 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 1.3 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 | 0.3 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 0.3 | GO:0098529 | neuromuscular junction development, skeletal muscle fiber(GO:0098529) |
| 0.1 | 0.9 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 0.3 | GO:0061551 | trigeminal ganglion development(GO:0061551) |
| 0.1 | 0.8 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.1 | 1.2 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 0.2 | GO:0055071 | detoxification of zinc ion(GO:0010312) manganese ion homeostasis(GO:0055071) stress response to zinc ion(GO:1990359) |
| 0.1 | 0.2 | GO:0042245 | RNA repair(GO:0042245) |
| 0.1 | 0.2 | GO:0070291 | N-acylethanolamine metabolic process(GO:0070291) |
| 0.1 | 0.4 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.1 | GO:0051230 | mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.1 | 0.4 | GO:0038026 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) reelin-mediated signaling pathway(GO:0038026) |
| 0.1 | 0.5 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 0.5 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.1 | 0.3 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.1 | 0.2 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.1 | 0.9 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.1 | 1.3 | GO:0060998 | regulation of dendritic spine development(GO:0060998) |
| 0.1 | 0.8 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.0 | 1.3 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 1.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.9 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.4 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.6 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 2.4 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.1 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.0 | 0.4 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.4 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.0 | 0.2 | GO:0003096 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.3 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 2.1 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 1.2 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.5 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.7 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 0.2 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.2 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.0 | 0.2 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 0.1 | GO:2000379 | positive regulation of reactive oxygen species metabolic process(GO:2000379) |
| 0.0 | 1.2 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 0.1 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.3 | GO:0003422 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.0 | 0.1 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.0 | 0.2 | GO:1900744 | regulation of p38MAPK cascade(GO:1900744) |
| 0.0 | 0.2 | GO:0042537 | benzene-containing compound metabolic process(GO:0042537) |
| 0.0 | 0.2 | GO:0099525 | synaptic vesicle docking(GO:0016081) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.0 | 0.1 | GO:2001204 | osteoclast development(GO:0036035) regulation of osteoclast development(GO:2001204) |
| 0.0 | 0.5 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.2 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.0 | 0.1 | GO:0060004 | reflex(GO:0060004) vestibular reflex(GO:0060005) |
| 0.0 | 0.5 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.1 | GO:0044406 | virion attachment to host cell(GO:0019062) adhesion of symbiont to host(GO:0044406) adhesion of symbiont to host cell(GO:0044650) |
| 0.0 | 0.2 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.0 | 0.4 | GO:0072576 | liver morphogenesis(GO:0072576) |
| 0.0 | 0.1 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.1 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.0 | 0.9 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.9 | GO:0055078 | sodium ion homeostasis(GO:0055078) |
| 0.0 | 1.4 | GO:0006171 | cAMP biosynthetic process(GO:0006171) |
| 0.0 | 0.9 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.3 | GO:0070828 | heterochromatin organization(GO:0070828) |
| 0.0 | 0.3 | GO:0033151 | V(D)J recombination(GO:0033151) |
| 0.0 | 0.1 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.1 | GO:0039529 | RIG-I signaling pathway(GO:0039529) regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039531) regulation of RIG-I signaling pathway(GO:0039535) |
| 0.0 | 0.5 | GO:0043506 | activation of JUN kinase activity(GO:0007257) regulation of JUN kinase activity(GO:0043506) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.2 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.0 | 0.1 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.3 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.2 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.0 | 0.4 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.1 | GO:1901881 | positive regulation of protein depolymerization(GO:1901881) regulation of barbed-end actin filament capping(GO:2000812) |
| 0.0 | 1.0 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.4 | GO:0046058 | cAMP catabolic process(GO:0006198) cAMP metabolic process(GO:0046058) |
| 0.0 | 0.4 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.3 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.0 | 1.3 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.4 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.9 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.1 | GO:0090266 | regulation of spindle checkpoint(GO:0090231) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.1 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.0 | 0.2 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.6 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.1 | GO:0035188 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.1 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.3 | GO:0070873 | regulation of glycogen metabolic process(GO:0070873) |
| 0.0 | 0.3 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.1 | GO:0044246 | regulation of collagen metabolic process(GO:0010712) regulation of collagen biosynthetic process(GO:0032965) regulation of multicellular organismal metabolic process(GO:0044246) |
| 0.0 | 0.1 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 | 0.3 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 1.3 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.2 | GO:0007035 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.0 | 0.2 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.1 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.0 | 0.0 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.6 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.6 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.0 | 1.3 | GO:0021782 | glial cell development(GO:0021782) |
| 0.0 | 0.0 | GO:0060347 | trabecula formation(GO:0060343) heart trabecula formation(GO:0060347) |
| 0.0 | 0.1 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.1 | GO:0009109 | coenzyme catabolic process(GO:0009109) |
| 0.0 | 0.4 | GO:0048599 | oocyte development(GO:0048599) |
| 0.0 | 1.5 | GO:0034976 | response to endoplasmic reticulum stress(GO:0034976) |
| 0.0 | 0.1 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.7 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0005948 | acetolactate synthase complex(GO:0005948) |
| 0.3 | 0.8 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.2 | 4.6 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.2 | 1.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.2 | 1.4 | GO:0030428 | cell septum(GO:0030428) |
| 0.1 | 0.7 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 0.4 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) anchored component of the cytoplasmic side of the plasma membrane(GO:0098753) |
| 0.1 | 1.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 1.0 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 1.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 0.6 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 0.4 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 0.3 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.1 | 0.3 | GO:0097519 | DNA recombinase complex(GO:0097519) |
| 0.1 | 1.2 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.1 | 0.6 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 0.4 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 0.4 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.9 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 2.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.8 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.3 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 1.8 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.5 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.2 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.4 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 2.8 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.2 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.4 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.7 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 1.9 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.1 | GO:0034359 | mature chylomicron(GO:0034359) |
| 0.0 | 0.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.1 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.0 | 0.1 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.5 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 1.2 | GO:0000786 | nucleosome(GO:0000786) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0003984 | acetolactate synthase activity(GO:0003984) |
| 0.3 | 1.0 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.3 | 2.5 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.3 | 0.9 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.3 | 0.8 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.3 | 1.3 | GO:0050780 | mu-type opioid receptor binding(GO:0031852) dopamine receptor binding(GO:0050780) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.2 | 0.7 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.2 | 0.7 | GO:0080132 | fatty acid alpha-hydroxylase activity(GO:0080132) |
| 0.2 | 0.6 | GO:0015228 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.2 | 1.6 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.2 | 1.4 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.2 | 1.4 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.2 | 0.8 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.1 | 1.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 1.4 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.1 | 0.7 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 0.6 | GO:2001070 | starch binding(GO:2001070) |
| 0.1 | 1.0 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 1.0 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.6 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 1.2 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 1.2 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.1 | 1.3 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 0.4 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 0.6 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.1 | 0.9 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 0.5 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 0.4 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.1 | 0.3 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.1 | 1.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.7 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 0.4 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 0.2 | GO:1990715 | mRNA CDS binding(GO:1990715) sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 1.1 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.3 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 2.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.4 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.5 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 0.3 | GO:1990238 | double-stranded DNA endodeoxyribonuclease activity(GO:1990238) |
| 0.1 | 0.3 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.1 | 0.6 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.1 | 0.9 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 0.2 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.1 | 0.2 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.1 | 0.9 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.7 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.6 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.1 | 0.8 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 1.4 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 0.6 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 0.3 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.5 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.6 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.0 | 0.7 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 1.1 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.5 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 1.8 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.2 | GO:1990931 | DNA-N1-methyladenine dioxygenase activity(GO:0043734) RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.0 | 0.8 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.3 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 2.7 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 1.0 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 1.9 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.3 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.3 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.3 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 8.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.8 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.5 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.4 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.6 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.2 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.4 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.4 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.2 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.9 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.0 | 0.6 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.5 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.3 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0090556 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 0.9 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 1.9 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 1.7 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.9 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.2 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.9 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.3 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.4 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.4 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.0 | 0.3 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.2 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.2 | GO:0008009 | chemokine activity(GO:0008009) chemokine receptor binding(GO:0042379) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 2.2 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.1 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.5 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.2 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.5 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.2 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.3 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 4.1 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.5 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.7 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.0 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.5 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 1.3 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 2.2 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.4 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.5 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.3 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.5 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.4 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.7 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 0.5 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 0.9 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 1.0 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 0.6 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 1.1 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 2.4 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.1 | 0.4 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.7 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.8 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.8 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.0 | 0.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.9 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.6 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 1.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.5 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.5 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.9 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.9 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.2 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.6 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.5 | REACTOME ION CHANNEL TRANSPORT | Genes involved in Ion channel transport |