Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
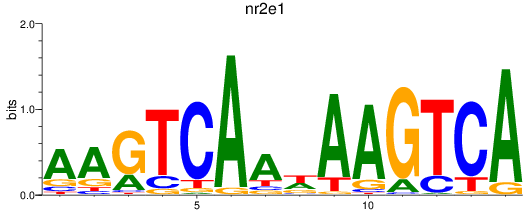
Results for nr2e1
Z-value: 0.63
Transcription factors associated with nr2e1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr2e1
|
ENSDARG00000017107 | nuclear receptor subfamily 2, group E, member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr2e1 | dr11_v1_chr20_-_32446406_32446406 | 0.33 | 1.6e-01 | Click! |
Activity profile of nr2e1 motif
Sorted Z-values of nr2e1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_9246018 | 1.49 |
ENSDART00000081769
ENSDART00000183189 |
susd1
|
sushi domain containing 1 |
| chr25_-_32381642 | 1.34 |
ENSDART00000133872
ENSDART00000006124 |
cops2
|
COP9 signalosome subunit 2 |
| chr1_-_33645967 | 1.27 |
ENSDART00000192758
|
cldng
|
claudin g |
| chr8_-_50287949 | 1.11 |
ENSDART00000023639
|
nkx2.7
|
NK2 transcription factor related 7 |
| chr22_+_22302614 | 0.89 |
ENSDART00000049434
|
scamp4
|
secretory carrier membrane protein 4 |
| chr23_+_44497701 | 0.88 |
ENSDART00000149903
|
si:ch73-375g18.1
|
si:ch73-375g18.1 |
| chr10_+_10351685 | 0.87 |
ENSDART00000109432
|
cercam
|
cerebral endothelial cell adhesion molecule |
| chr4_-_64482414 | 0.82 |
ENSDART00000159000
|
znf1105
|
zinc finger protein 1105 |
| chr13_-_35892051 | 0.81 |
ENSDART00000145884
|
tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr4_+_69577362 | 0.81 |
ENSDART00000160913
|
znf1030
|
zinc finger protein 1030 |
| chr5_-_50781623 | 0.78 |
ENSDART00000114950
|
zgc:194908
|
zgc:194908 |
| chr12_+_25945560 | 0.78 |
ENSDART00000109799
|
mmrn2b
|
multimerin 2b |
| chr7_+_26029672 | 0.76 |
ENSDART00000101126
|
alox12
|
arachidonate 12-lipoxygenase |
| chr22_-_22301929 | 0.74 |
ENSDART00000142027
|
chaf1a
|
chromatin assembly factor 1, subunit A (p150) |
| chr14_+_2487672 | 0.70 |
ENSDART00000170629
ENSDART00000123063 |
fgf18a
|
fibroblast growth factor 18a |
| chr20_+_30725778 | 0.69 |
ENSDART00000062532
|
nhsl1b
|
NHS-like 1b |
| chr10_-_35177257 | 0.69 |
ENSDART00000077426
|
pom121
|
POM121 transmembrane nucleoporin |
| chr13_-_35892243 | 0.68 |
ENSDART00000002750
ENSDART00000122810 ENSDART00000162399 |
tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr6_-_7769178 | 0.67 |
ENSDART00000191701
ENSDART00000149823 |
myh9a
|
myosin, heavy chain 9a, non-muscle |
| chr13_+_24671481 | 0.66 |
ENSDART00000001678
|
adam8a
|
ADAM metallopeptidase domain 8a |
| chr2_+_33926606 | 0.65 |
ENSDART00000111430
|
kif2c
|
kinesin family member 2C |
| chr22_-_22301672 | 0.65 |
ENSDART00000111711
|
chaf1a
|
chromatin assembly factor 1, subunit A (p150) |
| chr2_-_14272083 | 0.64 |
ENSDART00000169986
|
tctex1d1
|
Tctex1 domain containing 1 |
| chr23_+_28718863 | 0.64 |
ENSDART00000078156
|
srm
|
spermidine synthase |
| chr17_+_5985933 | 0.62 |
ENSDART00000190844
|
zgc:194275
|
zgc:194275 |
| chr2_+_10771787 | 0.62 |
ENSDART00000187782
|
gfi1aa
|
growth factor independent 1A transcription repressor a |
| chr7_-_38689562 | 0.61 |
ENSDART00000167209
|
aplnr2
|
apelin receptor 2 |
| chr12_-_26423439 | 0.60 |
ENSDART00000113978
|
synpo2lb
|
synaptopodin 2-like b |
| chr13_-_12660318 | 0.59 |
ENSDART00000008498
|
adh8a
|
alcohol dehydrogenase 8a |
| chr3_-_26017592 | 0.59 |
ENSDART00000030890
|
hmox1a
|
heme oxygenase 1a |
| chr3_-_26017831 | 0.58 |
ENSDART00000179982
|
hmox1a
|
heme oxygenase 1a |
| chr6_+_46406565 | 0.57 |
ENSDART00000168440
ENSDART00000131203 ENSDART00000138567 ENSDART00000132845 |
pbrm1l
|
polybromo 1, like |
| chr20_+_11731039 | 0.57 |
ENSDART00000152215
ENSDART00000152585 |
si:ch211-155o21.3
|
si:ch211-155o21.3 |
| chr18_-_5542351 | 0.57 |
ENSDART00000129459
|
spata5l1
|
spermatogenesis associated 5-like 1 |
| chr1_-_52292235 | 0.55 |
ENSDART00000132638
|
si:dkey-121b10.7
|
si:dkey-121b10.7 |
| chr4_+_5196469 | 0.55 |
ENSDART00000067386
|
rad51ap1
|
RAD51 associated protein 1 |
| chr1_-_9130136 | 0.55 |
ENSDART00000113013
|
palb2
|
partner and localizer of BRCA2 |
| chr13_-_6279684 | 0.54 |
ENSDART00000092105
|
agpat5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 (lysophosphatidic acid acyltransferase, epsilon) |
| chr17_+_28073062 | 0.53 |
ENSDART00000182021
|
AL844186.1
|
|
| chr20_+_33294428 | 0.51 |
ENSDART00000024104
|
mycn
|
MYCN proto-oncogene, bHLH transcription factor |
| chr23_-_16485190 | 0.50 |
ENSDART00000155038
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr4_-_77116266 | 0.50 |
ENSDART00000174249
|
CU467646.2
|
|
| chr4_-_71983381 | 0.50 |
ENSDART00000169130
|
ftr64
|
finTRIM family, member 64 |
| chr21_+_6394929 | 0.49 |
ENSDART00000138600
|
si:ch211-225g23.1
|
si:ch211-225g23.1 |
| chr13_-_15986871 | 0.49 |
ENSDART00000189394
|
ikzf1
|
IKAROS family zinc finger 1 (Ikaros) |
| chr4_-_8060962 | 0.48 |
ENSDART00000146622
|
wnk1b
|
WNK lysine deficient protein kinase 1b |
| chr21_+_25802190 | 0.48 |
ENSDART00000128987
|
nf2b
|
neurofibromin 2b (merlin) |
| chr4_+_59748607 | 0.48 |
ENSDART00000108499
|
znf1068
|
zinc finger protein 1068 |
| chr2_+_5843694 | 0.48 |
ENSDART00000182364
|
dis3l2
|
DIS3 like 3'-5' exoribonuclease 2 |
| chr4_-_59159690 | 0.47 |
ENSDART00000164706
|
znf1149
|
zinc finger protein 1149 |
| chr5_-_19861766 | 0.47 |
ENSDART00000088904
|
gltpa
|
glycolipid transfer protein a |
| chr14_-_26377044 | 0.47 |
ENSDART00000022236
|
emx3
|
empty spiracles homeobox 3 |
| chr4_+_43392655 | 0.46 |
ENSDART00000150797
|
znf1073
|
zinc finger protein 1073 |
| chr7_+_39416336 | 0.46 |
ENSDART00000171783
|
CT030188.1
|
|
| chr4_-_77125693 | 0.46 |
ENSDART00000174256
|
CU467646.3
|
|
| chr12_+_9551667 | 0.44 |
ENSDART00000048493
ENSDART00000166178 |
p4ha1a
|
prolyl 4-hydroxylase, alpha polypeptide I a |
| chr1_+_58608978 | 0.42 |
ENSDART00000152744
|
si:ch73-236c18.9
|
si:ch73-236c18.9 |
| chr19_-_3886991 | 0.41 |
ENSDART00000162085
|
thrap3b
|
thyroid hormone receptor associated protein 3b |
| chr16_+_10429770 | 0.41 |
ENSDART00000173132
|
vars
|
valyl-tRNA synthetase |
| chr8_+_23034718 | 0.41 |
ENSDART00000184512
|
ythdf1
|
YTH N(6)-methyladenosine RNA binding protein 1 |
| chr10_-_1625080 | 0.40 |
ENSDART00000137285
|
nup155
|
nucleoporin 155 |
| chr23_-_16484383 | 0.40 |
ENSDART00000187839
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr9_-_30220818 | 0.39 |
ENSDART00000140929
|
si:dkey-100n23.3
|
si:dkey-100n23.3 |
| chr5_+_28848870 | 0.39 |
ENSDART00000149563
|
zgc:174259
|
zgc:174259 |
| chr19_+_6914095 | 0.39 |
ENSDART00000104906
|
si:ch73-367f21.4
|
si:ch73-367f21.4 |
| chr6_-_43283122 | 0.39 |
ENSDART00000186022
|
frmd4ba
|
FERM domain containing 4Ba |
| chr2_-_48255843 | 0.38 |
ENSDART00000056277
|
gigyf2
|
GRB10 interacting GYF protein 2 |
| chr7_-_3669868 | 0.38 |
ENSDART00000172907
|
si:ch211-282j17.12
|
si:ch211-282j17.12 |
| chr22_+_13977772 | 0.37 |
ENSDART00000080313
|
arl4ca
|
ADP-ribosylation factor-like 4Ca |
| chr7_-_26571994 | 0.36 |
ENSDART00000128801
|
si:dkey-62k3.6
|
si:dkey-62k3.6 |
| chr6_+_58622831 | 0.35 |
ENSDART00000128793
|
sp7
|
Sp7 transcription factor |
| chr18_+_44631789 | 0.35 |
ENSDART00000144271
|
bicra
|
BRD4 interacting chromatin remodeling complex associated protein |
| chr13_+_24396666 | 0.33 |
ENSDART00000139197
ENSDART00000101200 |
zgc:153169
|
zgc:153169 |
| chr16_+_20915319 | 0.33 |
ENSDART00000079383
|
hoxa9b
|
homeobox A9b |
| chr24_+_35827766 | 0.33 |
ENSDART00000144700
|
si:dkeyp-7a3.1
|
si:dkeyp-7a3.1 |
| chr22_+_2409175 | 0.33 |
ENSDART00000141776
|
zgc:113220
|
zgc:113220 |
| chr5_+_13870340 | 0.33 |
ENSDART00000160690
|
hk2
|
hexokinase 2 |
| chr21_+_5129513 | 0.32 |
ENSDART00000102572
|
thbs4b
|
thrombospondin 4b |
| chr22_+_1837448 | 0.32 |
ENSDART00000160015
|
znf1183
|
zinc finger protein 1183 |
| chr5_+_61508418 | 0.32 |
ENSDART00000023676
|
bckdk
|
branched chain ketoacid dehydrogenase kinase |
| chr6_-_21534301 | 0.32 |
ENSDART00000126186
|
psmd12
|
proteasome 26S subunit, non-ATPase 12 |
| chr16_-_26855936 | 0.32 |
ENSDART00000167320
ENSDART00000078119 |
ino80c
|
INO80 complex subunit C |
| chr7_+_55112922 | 0.32 |
ENSDART00000073549
|
snai3
|
snail family zinc finger 3 |
| chr11_+_25508129 | 0.31 |
ENSDART00000011250
|
tfe3b
|
transcription factor binding to IGHM enhancer 3b |
| chr14_+_31865099 | 0.31 |
ENSDART00000189124
|
tm9sf5
|
transmembrane 9 superfamily protein member 5 |
| chr21_+_37411547 | 0.31 |
ENSDART00000076320
|
mrps17
|
mitochondrial ribosomal protein S17 |
| chr7_-_3693303 | 0.30 |
ENSDART00000166949
|
si:ch211-282j17.13
|
si:ch211-282j17.13 |
| chr15_-_4485828 | 0.30 |
ENSDART00000062868
|
tfdp2
|
transcription factor Dp-2 |
| chr4_-_72287764 | 0.29 |
ENSDART00000125452
ENSDART00000189437 |
si:cabz01071911.3
|
si:cabz01071911.3 |
| chr14_+_31865324 | 0.29 |
ENSDART00000039880
|
tm9sf5
|
transmembrane 9 superfamily protein member 5 |
| chr4_-_3064101 | 0.29 |
ENSDART00000135701
|
plekha5
|
pleckstrin homology domain containing, family A member 5 |
| chr15_-_33834577 | 0.28 |
ENSDART00000163354
|
mmp13b
|
matrix metallopeptidase 13b |
| chr22_-_6054749 | 0.28 |
ENSDART00000191603
|
CU468041.2
|
|
| chr2_+_18988407 | 0.28 |
ENSDART00000170216
|
glula
|
glutamate-ammonia ligase (glutamine synthase) a |
| chr15_+_37546391 | 0.28 |
ENSDART00000186625
|
psenen
|
presenilin enhancer gamma secretase subunit |
| chr5_-_48285756 | 0.28 |
ENSDART00000183495
|
mef2cb
|
myocyte enhancer factor 2cb |
| chr4_-_64542903 | 0.27 |
ENSDART00000143176
|
BX936382.1
|
|
| chr4_-_77120928 | 0.27 |
ENSDART00000174154
|
CU467646.1
|
|
| chr17_-_48743076 | 0.27 |
ENSDART00000173117
|
kcnk17
|
potassium channel, subfamily K, member 17 |
| chr10_+_8554929 | 0.27 |
ENSDART00000190849
|
tbc1d10ab
|
TBC1 domain family, member 10Ab |
| chr19_+_28284120 | 0.27 |
ENSDART00000103862
|
mrpl36
|
mitochondrial ribosomal protein L36 |
| chr4_-_59057046 | 0.26 |
ENSDART00000150249
|
znf1132
|
zinc finger protein 1132 |
| chr15_+_37545855 | 0.26 |
ENSDART00000099456
|
psenen
|
presenilin enhancer gamma secretase subunit |
| chr10_+_7593185 | 0.26 |
ENSDART00000162617
ENSDART00000162590 ENSDART00000171744 |
ppp2cb
|
protein phosphatase 2, catalytic subunit, beta isozyme |
| chr5_+_38886499 | 0.26 |
ENSDART00000076845
|
fras1
|
Fraser extracellular matrix complex subunit 1 |
| chr14_-_40389699 | 0.26 |
ENSDART00000181581
ENSDART00000173398 |
pcdh19
|
protocadherin 19 |
| chr23_-_333457 | 0.25 |
ENSDART00000114486
|
uhrf1bp1
|
UHRF1 binding protein 1 |
| chr7_+_52753067 | 0.25 |
ENSDART00000053814
|
mtfmt
|
mitochondrial methionyl-tRNA formyltransferase |
| chr4_+_60381608 | 0.25 |
ENSDART00000167351
|
znf1130
|
zinc finger protein 1130 |
| chr10_-_15644904 | 0.24 |
ENSDART00000138389
ENSDART00000101191 ENSDART00000186559 ENSDART00000122170 |
smc5
|
structural maintenance of chromosomes 5 |
| chr2_-_55861351 | 0.24 |
ENSDART00000059003
|
rx2
|
retinal homeobox gene 2 |
| chr1_-_51266394 | 0.24 |
ENSDART00000164016
|
kif16ba
|
kinesin family member 16Ba |
| chr21_-_25801956 | 0.23 |
ENSDART00000101219
|
mettl27
|
methyltransferase like 27 |
| chr8_+_30619919 | 0.22 |
ENSDART00000014046
|
specc1la
|
sperm antigen with calponin homology and coiled-coil domains 1-like a |
| chr4_-_43071336 | 0.22 |
ENSDART00000150773
|
znf1072
|
zinc finger protein 1072 |
| chr21_+_20356458 | 0.22 |
ENSDART00000135816
|
pde6b
|
phosphodiesterase 6B, cGMP-specific, rod, beta |
| chr2_+_30249977 | 0.21 |
ENSDART00000109160
ENSDART00000135171 |
tmem70
|
transmembrane protein 70 |
| chr12_-_23128746 | 0.21 |
ENSDART00000170018
|
armc4
|
armadillo repeat containing 4 |
| chr16_+_38360002 | 0.21 |
ENSDART00000087346
ENSDART00000148101 |
zgc:113232
|
zgc:113232 |
| chr6_+_46668717 | 0.21 |
ENSDART00000064853
|
oxtrl
|
oxytocin receptor like |
| chr21_+_19834072 | 0.21 |
ENSDART00000147555
|
ccdc80l2
|
coiled-coil domain containing 80 like 2 |
| chr5_-_63663811 | 0.21 |
ENSDART00000111403
|
ciz1b
|
cdkn1a interacting zinc finger protein 1b |
| chr25_-_15507523 | 0.20 |
ENSDART00000134166
|
si:dkeyp-67e1.3
|
si:dkeyp-67e1.3 |
| chr18_-_18662577 | 0.20 |
ENSDART00000076695
|
aars
|
alanyl-tRNA synthetase |
| chr17_-_49581414 | 0.20 |
ENSDART00000106339
|
CU929037.1
|
|
| chr1_+_14073891 | 0.20 |
ENSDART00000021693
|
ank2a
|
ankyrin 2a, neuronal |
| chr11_+_19433936 | 0.20 |
ENSDART00000162081
|
prickle2b
|
prickle homolog 2b |
| chr17_+_33158350 | 0.19 |
ENSDART00000104476
|
snx9a
|
sorting nexin 9a |
| chr8_-_39884359 | 0.19 |
ENSDART00000131372
|
mlec
|
malectin |
| chr7_-_30367650 | 0.19 |
ENSDART00000075519
|
aldh1a2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr2_-_29923630 | 0.19 |
ENSDART00000158844
ENSDART00000031130 |
paxip1
|
PAX interacting (with transcription-activation domain) protein 1 |
| chr20_-_25877793 | 0.18 |
ENSDART00000177333
ENSDART00000184619 |
exoc1
|
exocyst complex component 1 |
| chr19_-_10286228 | 0.18 |
ENSDART00000027316
|
cnot3b
|
CCR4-NOT transcription complex, subunit 3b |
| chr21_-_28901095 | 0.18 |
ENSDART00000180820
|
cxxc5a
|
CXXC finger protein 5a |
| chr24_+_20658942 | 0.18 |
ENSDART00000142848
|
sec22c
|
SEC22 homolog C, vesicle trafficking protein |
| chr7_+_4648398 | 0.17 |
ENSDART00000148099
|
si:dkey-83f18.3
|
si:dkey-83f18.3 |
| chr20_+_30578163 | 0.17 |
ENSDART00000159485
|
fgfr1op
|
FGFR1 oncogene partner |
| chr20_+_47196506 | 0.16 |
ENSDART00000153040
|
znf975
|
zinc finger protein 975 |
| chr13_-_24396199 | 0.15 |
ENSDART00000181093
|
tbp
|
TATA box binding protein |
| chr6_-_40435025 | 0.15 |
ENSDART00000031153
|
ccdc174
|
coiled-coil domain containing 174 |
| chr7_+_57108823 | 0.15 |
ENSDART00000184943
ENSDART00000055956 |
enosf1
|
enolase superfamily member 1 |
| chr23_-_14918276 | 0.14 |
ENSDART00000179831
|
ndrg3b
|
ndrg family member 3b |
| chr23_+_39970425 | 0.14 |
ENSDART00000165376
|
fyco1a
|
FYVE and coiled-coil domain containing 1a |
| chr6_-_49655879 | 0.14 |
ENSDART00000191594
|
apcdd1l
|
adenomatosis polyposis coli down-regulated 1-like |
| chr7_+_38770167 | 0.13 |
ENSDART00000190827
|
arhgap1
|
Rho GTPase activating protein 1 |
| chr12_+_7865470 | 0.13 |
ENSDART00000161683
|
BX548028.1
|
|
| chr9_+_16854121 | 0.13 |
ENSDART00000110866
|
cln5
|
CLN5, intracellular trafficking protein |
| chr22_-_17652112 | 0.13 |
ENSDART00000189205
|
hmha1b
|
histocompatibility (minor) HA-1 b |
| chr24_+_20658760 | 0.13 |
ENSDART00000188362
|
sec22c
|
SEC22 homolog C, vesicle trafficking protein |
| chr4_-_37708661 | 0.13 |
ENSDART00000168788
|
znf1139
|
zinc finger protein 1139 |
| chr7_+_59033902 | 0.11 |
ENSDART00000168888
|
fastkd3
|
FAST kinase domains 3 |
| chr11_-_26576754 | 0.11 |
ENSDART00000191733
ENSDART00000002846 |
st3gal8
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 8 |
| chr9_+_38369872 | 0.11 |
ENSDART00000193914
|
plcd4b
|
phospholipase C, delta 4b |
| chr1_-_46244523 | 0.10 |
ENSDART00000143908
|
si:ch211-138g9.3
|
si:ch211-138g9.3 |
| chr17_-_12408109 | 0.10 |
ENSDART00000155509
|
ankef1b
|
ankyrin repeat and EF-hand domain containing 1b |
| chr4_+_37239091 | 0.10 |
ENSDART00000172013
|
znf973
|
zinc finger protein 973 |
| chr6_-_37744430 | 0.10 |
ENSDART00000150177
ENSDART00000149722 |
nipa2
|
non imprinted in Prader-Willi/Angelman syndrome 2 (human) |
| chr6_-_52723901 | 0.10 |
ENSDART00000033949
|
oser1
|
oxidative stress responsive serine-rich 1 |
| chr25_-_16160494 | 0.09 |
ENSDART00000146880
|
si:dkey-80c24.4
|
si:dkey-80c24.4 |
| chr16_+_33111947 | 0.08 |
ENSDART00000058473
|
cnr2
|
cannabinoid receptor 2 |
| chr5_+_43782267 | 0.08 |
ENSDART00000130355
|
nos2a
|
nitric oxide synthase 2a, inducible |
| chr21_+_5508167 | 0.08 |
ENSDART00000157702
|
ly6m4
|
lymphocyte antigen 6 family member M4 |
| chr4_+_71735828 | 0.07 |
ENSDART00000174867
|
znf1116
|
zinc finger protein 1116 |
| chr1_+_44003654 | 0.07 |
ENSDART00000131337
|
si:dkey-22i16.10
|
si:dkey-22i16.10 |
| chr13_+_8958438 | 0.06 |
ENSDART00000165721
ENSDART00000169398 |
haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr4_-_2868112 | 0.06 |
ENSDART00000133843
|
pde3a
|
phosphodiesterase 3A, cGMP-inhibited |
| chr4_+_76926059 | 0.06 |
ENSDART00000136192
|
si:dkey-240n22.6
|
si:dkey-240n22.6 |
| chr14_-_45604667 | 0.06 |
ENSDART00000167016
|
si:ch211-276i12.9
|
si:ch211-276i12.9 |
| chr17_+_29345606 | 0.06 |
ENSDART00000086164
|
kctd3
|
potassium channel tetramerization domain containing 3 |
| chr2_+_21486529 | 0.05 |
ENSDART00000047468
|
inhbab
|
inhibin, beta Ab |
| chr22_+_28969071 | 0.05 |
ENSDART00000163427
|
pimr95
|
Pim proto-oncogene, serine/threonine kinase, related 95 |
| chr4_-_39942347 | 0.05 |
ENSDART00000124410
|
znf979
|
zinc finger protein 979 |
| chr24_+_28614576 | 0.04 |
ENSDART00000184941
|
abca4a
|
ATP-binding cassette, sub-family A (ABC1), member 4a |
| chr21_+_21791799 | 0.04 |
ENSDART00000151759
|
neu3.1
|
sialidase 3 (membrane sialidase), tandem duplicate 1 |
| chr9_-_31135901 | 0.04 |
ENSDART00000113027
ENSDART00000193210 ENSDART00000128896 |
si:ch211-184m13.4
|
si:ch211-184m13.4 |
| chr14_+_44774089 | 0.04 |
ENSDART00000098643
|
tomm5
|
translocase of outer mitochondrial membrane 5 homolog (yeast) |
| chr23_-_19682971 | 0.03 |
ENSDART00000048891
|
zgc:193598
|
zgc:193598 |
| chr17_+_14784275 | 0.03 |
ENSDART00000005355
|
rtraf
|
RNA transcription, translation and transport factor |
| chr5_-_45877387 | 0.03 |
ENSDART00000183714
ENSDART00000041503 |
slc4a4a
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4a |
| chr22_-_24880824 | 0.03 |
ENSDART00000061165
|
vtg2
|
vitellogenin 2 |
| chr16_+_28932038 | 0.02 |
ENSDART00000149480
|
npr1b
|
natriuretic peptide receptor 1b |
| chr21_+_28535203 | 0.02 |
ENSDART00000184950
|
CR749775.2
|
|
| chr18_+_31016379 | 0.02 |
ENSDART00000172461
ENSDART00000163471 |
uraha
|
urate (5-hydroxyiso-) hydrolase a |
| chr2_-_20890429 | 0.02 |
ENSDART00000149989
|
pdca
|
phosducin a |
| chr7_-_59047720 | 0.02 |
ENSDART00000184831
|
CR376779.1
|
|
| chr25_-_31433512 | 0.01 |
ENSDART00000067028
|
zgc:172145
|
zgc:172145 |
| chr16_-_39178979 | 0.01 |
ENSDART00000158894
|
si:dkey-99n20.6
|
si:dkey-99n20.6 |
| chr16_-_35392926 | 0.01 |
ENSDART00000164482
|
si:dkey-34d22.3
|
si:dkey-34d22.3 |
| chr7_-_41693004 | 0.00 |
ENSDART00000121509
|
malrd1
|
MAM and LDL receptor class A domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr2e1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0007585 | respiratory gaseous exchange(GO:0007585) regulation of respiratory gaseous exchange(GO:0043576) |
| 0.3 | 1.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.3 | 1.3 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.2 | 0.7 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.4 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) protein localization to nuclear envelope(GO:0090435) |
| 0.1 | 0.7 | GO:0072103 | renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.1 | 0.5 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.1 | 0.4 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.1 | 1.1 | GO:0003207 | cardiac chamber formation(GO:0003207) |
| 0.1 | 0.6 | GO:0046294 | formaldehyde metabolic process(GO:0046292) formaldehyde catabolic process(GO:0046294) |
| 0.1 | 0.3 | GO:0035989 | tendon development(GO:0035989) |
| 0.1 | 0.2 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.1 | 0.6 | GO:0008216 | spermidine metabolic process(GO:0008216) |
| 0.1 | 0.2 | GO:0007567 | parturition(GO:0007567) maternal process involved in parturition(GO:0060137) |
| 0.1 | 0.8 | GO:0051122 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 0.5 | GO:0034205 | Notch receptor processing(GO:0007220) beta-amyloid formation(GO:0034205) amyloid precursor protein catabolic process(GO:0042987) |
| 0.1 | 0.4 | GO:0009146 | dGTP catabolic process(GO:0006203) purine nucleoside triphosphate catabolic process(GO:0009146) purine deoxyribonucleoside triphosphate catabolic process(GO:0009217) dGTP metabolic process(GO:0046070) |
| 0.1 | 0.7 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.2 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.3 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.7 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.4 | GO:0050779 | RNA destabilization(GO:0050779) mRNA destabilization(GO:0061157) |
| 0.0 | 0.3 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.3 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.5 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.1 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.5 | GO:0033077 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.0 | 0.4 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.3 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.4 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.3 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.0 | 0.6 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.5 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.2 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.0 | 0.6 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.5 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.2 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.1 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.0 | 1.4 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.1 | GO:0097241 | hematopoietic stem cell migration to bone marrow(GO:0097241) |
| 0.0 | 0.7 | GO:0006405 | RNA export from nucleus(GO:0006405) |
| 0.0 | 0.1 | GO:0031282 | regulation of cGMP metabolic process(GO:0030823) positive regulation of cGMP metabolic process(GO:0030825) regulation of cGMP biosynthetic process(GO:0030826) positive regulation of cGMP biosynthetic process(GO:0030828) regulation of guanylate cyclase activity(GO:0031282) positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 1.0 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.0 | 0.1 | GO:0043420 | anthranilate metabolic process(GO:0043420) |
| 0.0 | 0.6 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.4 | GO:0017121 | phospholipid scrambling(GO:0017121) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.2 | 0.7 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.1 | 1.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 0.4 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 0.6 | GO:0016586 | RSC complex(GO:0016586) |
| 0.1 | 0.5 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.2 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.5 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 1.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.3 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.3 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.7 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.3 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.2 | 0.9 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.2 | 1.2 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.6 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.1 | 0.6 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) S-(hydroxymethyl)glutathione dehydrogenase activity(GO:0051903) |
| 0.1 | 0.3 | GO:0005252 | open rectifier potassium channel activity(GO:0005252) |
| 0.1 | 0.7 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 0.4 | GO:0016793 | dGTPase activity(GO:0008832) triphosphoric monoester hydrolase activity(GO:0016793) |
| 0.1 | 0.2 | GO:0004990 | oxytocin receptor activity(GO:0004990) |
| 0.1 | 0.4 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.5 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.3 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.4 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.7 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.3 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.3 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 1.1 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.4 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.5 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.4 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.2 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.0 | 0.2 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 0.2 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.8 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 1.8 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.5 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.5 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.2 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.5 | PID FANCONI PATHWAY | Fanconi anemia pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.0 | 0.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.6 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 1.4 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.5 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |