Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
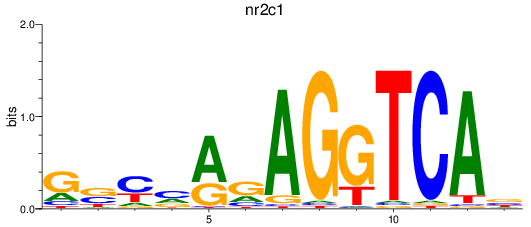
Results for nr2c1
Z-value: 0.49
Transcription factors associated with nr2c1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr2c1
|
ENSDARG00000045527 | nuclear receptor subfamily 2, group C, member 1 |
|
nr2c1
|
ENSDARG00000111803 | nuclear receptor subfamily 2, group C, member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr2c1 | dr11_v1_chr4_-_25858620_25858620 | -0.73 | 3.7e-04 | Click! |
Activity profile of nr2c1 motif
Sorted Z-values of nr2c1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_26017360 | 2.35 |
ENSDART00000149466
|
prss59.2
|
protease, serine, 59, tandem duplicate 2 |
| chr10_-_44027391 | 2.31 |
ENSDART00000145404
|
crybb1
|
crystallin, beta B1 |
| chr16_+_26012569 | 2.30 |
ENSDART00000148846
|
prss59.1
|
protease, serine, 59, tandem duplicate 1 |
| chr15_+_19797918 | 1.73 |
ENSDART00000113314
|
si:ch211-229d2.5
|
si:ch211-229d2.5 |
| chr23_-_39636195 | 1.42 |
ENSDART00000144439
|
vwa1
|
von Willebrand factor A domain containing 1 |
| chr7_+_73332486 | 1.41 |
ENSDART00000174119
ENSDART00000092051 ENSDART00000192388 |
CABZ01081780.1
|
|
| chr2_+_38039857 | 1.39 |
ENSDART00000159951
|
casq1a
|
calsequestrin 1a |
| chr23_-_45504991 | 1.33 |
ENSDART00000148761
|
col24a1
|
collagen type XXIV alpha 1 |
| chr13_-_303137 | 1.21 |
ENSDART00000099131
|
chs1
|
chitin synthase 1 |
| chr5_-_23280098 | 1.20 |
ENSDART00000126540
ENSDART00000051533 |
plp1b
|
proteolipid protein 1b |
| chr11_+_36989696 | 1.11 |
ENSDART00000045888
|
tkta
|
transketolase a |
| chr4_-_4387012 | 1.09 |
ENSDART00000191836
|
CU468826.3
|
Danio rerio U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit-related protein 1-like (LOC100331497), mRNA. |
| chr15_+_47386939 | 1.09 |
ENSDART00000128224
|
FO904873.1
|
|
| chr7_-_20756013 | 1.01 |
ENSDART00000185259
|
chd3
|
chromodomain helicase DNA binding protein 3 |
| chr17_+_12408405 | 1.00 |
ENSDART00000154827
ENSDART00000048440 ENSDART00000156429 |
khk
|
ketohexokinase |
| chr19_-_3193912 | 0.96 |
ENSDART00000133159
|
si:ch211-133n4.6
|
si:ch211-133n4.6 |
| chr1_-_19215336 | 0.95 |
ENSDART00000162949
ENSDART00000170680 |
ptprdb
|
protein tyrosine phosphatase, receptor type, D, b |
| chr3_+_30257582 | 0.94 |
ENSDART00000159497
ENSDART00000103457 ENSDART00000121883 |
mybpc2a
|
myosin binding protein C, fast type a |
| chr17_+_12408188 | 0.87 |
ENSDART00000105218
|
khk
|
ketohexokinase |
| chr7_-_26457208 | 0.87 |
ENSDART00000173519
|
zgc:172079
|
zgc:172079 |
| chr25_-_19420949 | 0.87 |
ENSDART00000181338
|
map1ab
|
microtubule-associated protein 1Ab |
| chr19_+_40861853 | 0.83 |
ENSDART00000126470
|
zgc:85777
|
zgc:85777 |
| chr23_+_216012 | 0.81 |
ENSDART00000181115
ENSDART00000004678 ENSDART00000190439 ENSDART00000189322 |
PDZD4
|
si:ch73-162j3.4 |
| chr24_+_39137001 | 0.79 |
ENSDART00000181086
ENSDART00000183724 ENSDART00000193466 |
tbc1d24
|
TBC1 domain family, member 24 |
| chr25_+_14092871 | 0.78 |
ENSDART00000067239
|
guca1g
|
guanylate cyclase activator 1g |
| chr24_-_38574631 | 0.76 |
ENSDART00000154539
|
slc17a7b
|
solute carrier family 17 (vesicular glutamate transporter), member 7b |
| chr8_-_12468744 | 0.75 |
ENSDART00000135019
|
FIBCD1 (1 of many)
|
si:dkeyp-51b7.3 |
| chr11_-_17713987 | 0.73 |
ENSDART00000090401
|
fam19a4b
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A4b |
| chr19_-_3193443 | 0.72 |
ENSDART00000179855
|
si:ch211-133n4.6
|
si:ch211-133n4.6 |
| chr22_-_38543630 | 0.70 |
ENSDART00000172029
|
si:ch211-126j24.1
|
si:ch211-126j24.1 |
| chr23_+_37482727 | 0.69 |
ENSDART00000162737
|
agmat
|
agmatine ureohydrolase (agmatinase) |
| chr11_+_30729745 | 0.65 |
ENSDART00000103270
|
slc22a7a
|
solute carrier family 22 (organic anion transporter), member 7a |
| chr8_-_11146924 | 0.64 |
ENSDART00000126824
|
asb13b
|
ankyrin repeat and SOCS box containing 13b |
| chr20_-_36671660 | 0.63 |
ENSDART00000134819
|
slc5a6a
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6a |
| chr18_-_15771551 | 0.61 |
ENSDART00000130931
ENSDART00000154079 |
si:ch211-219a15.3
|
si:ch211-219a15.3 |
| chr15_-_28618502 | 0.59 |
ENSDART00000086902
|
slc6a4a
|
solute carrier family 6 (neurotransmitter transporter), member 4a |
| chr5_+_31049742 | 0.58 |
ENSDART00000173097
|
zzef1
|
zinc finger, ZZ-type with EF hand domain 1 |
| chr23_+_33963619 | 0.57 |
ENSDART00000140666
ENSDART00000084792 |
plpbp
|
pyridoxal phosphate binding protein |
| chr2_-_53925308 | 0.57 |
ENSDART00000190705
|
FO834855.1
|
|
| chr5_-_68927728 | 0.57 |
ENSDART00000132838
|
ank1a
|
ankyrin 1, erythrocytic a |
| chr16_-_2558653 | 0.54 |
ENSDART00000110365
|
adcy3a
|
adenylate cyclase 3a |
| chr11_+_24819624 | 0.52 |
ENSDART00000155514
|
kdm5ba
|
lysine (K)-specific demethylase 5Ba |
| chr16_-_45209684 | 0.52 |
ENSDART00000184595
|
fxyd1
|
FXYD domain containing ion transport regulator 1 (phospholemman) |
| chr24_-_29586082 | 0.51 |
ENSDART00000136763
|
vav3a
|
vav 3 guanine nucleotide exchange factor a |
| chr1_-_46343999 | 0.50 |
ENSDART00000145117
ENSDART00000193233 |
atp11a
|
ATPase phospholipid transporting 11A |
| chr11_+_77526 | 0.49 |
ENSDART00000193521
|
CABZ01072242.1
|
|
| chr22_+_20427170 | 0.49 |
ENSDART00000136744
|
foxq2
|
forkhead box Q2 |
| chr2_-_57076687 | 0.49 |
ENSDART00000161523
|
slc25a42
|
solute carrier family 25, member 42 |
| chr23_+_4583747 | 0.48 |
ENSDART00000160350
|
LO017700.1
|
|
| chr7_-_42206720 | 0.48 |
ENSDART00000110907
|
itfg1
|
integrin alpha FG-GAP repeat containing 1 |
| chr22_-_7129631 | 0.48 |
ENSDART00000171359
|
asic1b
|
acid-sensing (proton-gated) ion channel 1b |
| chr21_-_1972236 | 0.46 |
ENSDART00000192858
ENSDART00000189962 ENSDART00000182461 ENSDART00000165547 |
WDR7
|
WD repeat domain 7 |
| chr1_-_46859398 | 0.45 |
ENSDART00000135795
|
agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr1_-_22726233 | 0.45 |
ENSDART00000140920
|
prom1b
|
prominin 1 b |
| chr16_+_30002605 | 0.45 |
ENSDART00000160555
|
sema6e
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6E |
| chr2_-_38141997 | 0.45 |
ENSDART00000143469
|
trim110
|
tripartite motif containing 110 |
| chr21_+_1119046 | 0.45 |
ENSDART00000184678
|
CABZ01088049.1
|
|
| chr7_+_50109239 | 0.45 |
ENSDART00000021605
|
LRRC4C (1 of many)
|
si:dkey-6l15.1 |
| chr14_-_27121854 | 0.45 |
ENSDART00000173119
|
pcdh11
|
protocadherin 11 |
| chr12_-_15620090 | 0.44 |
ENSDART00000038032
|
acbd4
|
acyl-CoA binding domain containing 4 |
| chr10_-_43404027 | 0.43 |
ENSDART00000086227
|
edil3b
|
EGF-like repeats and discoidin I-like domains 3b |
| chr3_-_58455289 | 0.43 |
ENSDART00000052179
|
cdr2a
|
cerebellar degeneration-related protein 2a |
| chr18_+_27515640 | 0.42 |
ENSDART00000181593
|
tp53i11b
|
tumor protein p53 inducible protein 11b |
| chr7_+_42206847 | 0.42 |
ENSDART00000149250
|
phkb
|
phosphorylase kinase, beta |
| chr2_+_42191592 | 0.41 |
ENSDART00000144716
|
cavin4a
|
caveolae associated protein 4a |
| chr11_+_11201096 | 0.41 |
ENSDART00000171916
ENSDART00000171521 ENSDART00000087105 ENSDART00000159603 |
myom2a
|
myomesin 2a |
| chr3_-_60308937 | 0.41 |
ENSDART00000136816
|
si:ch211-214b16.3
|
si:ch211-214b16.3 |
| chr3_+_54047342 | 0.41 |
ENSDART00000178486
|
olfm2a
|
olfactomedin 2a |
| chr24_-_21674950 | 0.40 |
ENSDART00000123216
ENSDART00000046211 |
lnx2a
|
ligand of numb-protein X 2a |
| chr3_-_34724879 | 0.40 |
ENSDART00000177021
|
thraa
|
thyroid hormone receptor alpha a |
| chr6_+_154556 | 0.39 |
ENSDART00000193153
|
zglp1
|
zinc finger, GATA-like protein 1 |
| chr10_+_42374770 | 0.39 |
ENSDART00000020000
|
COX5B
|
zgc:86599 |
| chr6_+_9872261 | 0.38 |
ENSDART00000045070
|
MPP4 (1 of many)
|
si:ch211-222n4.6 |
| chr7_-_29231686 | 0.38 |
ENSDART00000173780
|
herc1
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
| chr3_-_30609659 | 0.38 |
ENSDART00000182516
ENSDART00000187047 ENSDART00000110597 |
syt3
|
synaptotagmin III |
| chr10_-_322769 | 0.37 |
ENSDART00000165244
|
akt2l
|
v-akt murine thymoma viral oncogene homolog 2, like |
| chr7_+_34297271 | 0.37 |
ENSDART00000180342
|
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr23_-_46126444 | 0.37 |
ENSDART00000030004
|
CABZ01071903.1
|
|
| chr11_-_36020005 | 0.36 |
ENSDART00000031993
|
itpr1b
|
inositol 1,4,5-trisphosphate receptor, type 1b |
| chr24_-_31942656 | 0.36 |
ENSDART00000180308
ENSDART00000016879 |
c1ql3a
|
complement component 1, q subcomponent-like 3a |
| chr5_-_10768258 | 0.35 |
ENSDART00000157043
|
rtn4r
|
reticulon 4 receptor |
| chr24_-_31425799 | 0.35 |
ENSDART00000157998
|
cngb3.1
|
cyclic nucleotide gated channel beta 3, tandem duplicate 1 |
| chr10_-_29900546 | 0.35 |
ENSDART00000147441
|
lim2.1
|
lens intrinsic membrane protein 2.1 |
| chr14_+_1719367 | 0.35 |
ENSDART00000157696
|
trpc7b
|
transient receptor potential cation channel, subfamily C, member 7b |
| chr23_-_35069805 | 0.34 |
ENSDART00000087219
|
BX294434.1
|
|
| chr25_-_35113891 | 0.34 |
ENSDART00000190724
|
zgc:165555
|
zgc:165555 |
| chr2_-_4787566 | 0.34 |
ENSDART00000160663
ENSDART00000157808 |
tnk2b
|
tyrosine kinase, non-receptor, 2b |
| chr19_-_25519612 | 0.34 |
ENSDART00000133150
|
C1GALT1 (1 of many)
|
si:dkey-202e17.1 |
| chr8_-_4097722 | 0.34 |
ENSDART00000135006
|
cux2b
|
cut-like homeobox 2b |
| chr11_+_36243774 | 0.34 |
ENSDART00000023323
|
zgc:172270
|
zgc:172270 |
| chr10_-_40939706 | 0.32 |
ENSDART00000059795
ENSDART00000190510 |
bmp1b
|
bone morphogenetic protein 1b |
| chr3_-_23474416 | 0.32 |
ENSDART00000161672
|
si:dkey-225f5.5
|
si:dkey-225f5.5 |
| chr13_+_42406883 | 0.31 |
ENSDART00000084354
|
cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr12_-_44199316 | 0.31 |
ENSDART00000170378
|
si:ch73-329n5.1
|
si:ch73-329n5.1 |
| chr23_+_27912079 | 0.30 |
ENSDART00000171859
|
BX539336.1
|
|
| chr8_+_36948256 | 0.30 |
ENSDART00000140410
|
iqsec2b
|
IQ motif and Sec7 domain 2b |
| chr13_-_32577386 | 0.30 |
ENSDART00000016535
|
kcns3a
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3a |
| chr23_+_39970425 | 0.29 |
ENSDART00000165376
|
fyco1a
|
FYVE and coiled-coil domain containing 1a |
| chr13_+_13805385 | 0.29 |
ENSDART00000166859
|
slc4a11
|
solute carrier family 4, sodium borate transporter, member 11 |
| chr6_+_27339962 | 0.29 |
ENSDART00000193726
|
klhl30
|
kelch-like family member 30 |
| chr6_-_42112191 | 0.29 |
ENSDART00000085472
|
grm2a
|
glutamate receptor, metabotropic 2a |
| chr21_-_45728069 | 0.28 |
ENSDART00000185101
|
CU302316.1
|
|
| chr7_-_38792543 | 0.28 |
ENSDART00000157416
|
si:dkey-23n7.10
|
si:dkey-23n7.10 |
| chr10_-_40939303 | 0.27 |
ENSDART00000134295
|
bmp1b
|
bone morphogenetic protein 1b |
| chr17_-_49995366 | 0.27 |
ENSDART00000075223
|
cox7a2a
|
cytochrome c oxidase subunit VIIa polypeptide 2a (liver) |
| chr13_+_28819768 | 0.27 |
ENSDART00000191401
ENSDART00000188895 ENSDART00000101653 |
CU639469.1
|
|
| chr6_-_42111937 | 0.26 |
ENSDART00000181772
|
grm2a
|
glutamate receptor, metabotropic 2a |
| chr1_+_44582369 | 0.26 |
ENSDART00000003022
ENSDART00000137980 |
med19b
|
mediator complex subunit 19b |
| chr17_-_47305034 | 0.25 |
ENSDART00000150116
|
alk
|
ALK receptor tyrosine kinase |
| chr23_+_45611980 | 0.25 |
ENSDART00000181582
|
dclk2b
|
doublecortin-like kinase 2b |
| chr7_+_14005111 | 0.24 |
ENSDART00000187365
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr15_+_28119284 | 0.23 |
ENSDART00000188366
|
unc119a
|
unc-119 homolog a (C. elegans) |
| chr3_-_32927516 | 0.23 |
ENSDART00000140117
|
aoc2
|
amine oxidase, copper containing 2 |
| chr15_+_28268135 | 0.22 |
ENSDART00000152536
ENSDART00000188550 |
myo1cb
|
myosin Ic, paralog b |
| chr17_+_450956 | 0.21 |
ENSDART00000183022
ENSDART00000171386 |
zgc:194887
|
zgc:194887 |
| chr21_+_15883546 | 0.21 |
ENSDART00000186325
|
fam169ab
|
family with sequence similarity 169, member Ab |
| chr1_+_58182400 | 0.20 |
ENSDART00000144784
|
si:ch211-15j1.1
|
si:ch211-15j1.1 |
| chr17_-_5583345 | 0.19 |
ENSDART00000035944
|
clic5a
|
chloride intracellular channel 5a |
| chr1_+_58221372 | 0.19 |
ENSDART00000138467
|
si:dkey-222h21.10
|
si:dkey-222h21.10 |
| chr9_+_21151138 | 0.19 |
ENSDART00000133903
|
hao2
|
hydroxyacid oxidase 2 (long chain) |
| chr9_+_34433053 | 0.19 |
ENSDART00000137416
|
si:ch211-269e2.1
|
si:ch211-269e2.1 |
| chr6_+_40952031 | 0.18 |
ENSDART00000189219
|
patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr11_+_26609110 | 0.18 |
ENSDART00000042322
|
map1lc3a
|
microtubule-associated protein 1 light chain 3 alpha |
| chr2_+_43755274 | 0.18 |
ENSDART00000138620
|
arhgap12a
|
Rho GTPase activating protein 12a |
| chr8_-_979735 | 0.17 |
ENSDART00000149612
|
znf366
|
zinc finger protein 366 |
| chr14_+_22293787 | 0.17 |
ENSDART00000157967
|
atp10b
|
ATPase phospholipid transporting 10B |
| chr12_-_47899497 | 0.17 |
ENSDART00000162219
|
QRFPR
|
pyroglutamylated RFamide peptide receptor |
| chr10_+_42374957 | 0.16 |
ENSDART00000147926
|
COX5B
|
zgc:86599 |
| chr1_-_53750522 | 0.16 |
ENSDART00000190755
|
akt3b
|
v-akt murine thymoma viral oncogene homolog 3b |
| chr21_+_43171013 | 0.15 |
ENSDART00000151362
|
zcchc10
|
zinc finger, CCHC domain containing 10 |
| chr13_+_7575563 | 0.15 |
ENSDART00000146592
|
gbf1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr13_-_12581388 | 0.15 |
ENSDART00000079655
|
enpep
|
glutamyl aminopeptidase |
| chr23_-_2448234 | 0.15 |
ENSDART00000082097
|
LO018513.1
|
|
| chr5_-_66301142 | 0.15 |
ENSDART00000067541
|
prlrb
|
prolactin receptor b |
| chr1_+_58353661 | 0.14 |
ENSDART00000140074
|
si:dkey-222h21.2
|
si:dkey-222h21.2 |
| chr19_-_25519310 | 0.14 |
ENSDART00000089882
|
C1GALT1 (1 of many)
|
si:dkey-202e17.1 |
| chr8_-_11146687 | 0.14 |
ENSDART00000189626
|
asb13b
|
ankyrin repeat and SOCS box containing 13b |
| chr22_-_15280638 | 0.14 |
ENSDART00000063008
|
mfng
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr16_-_560574 | 0.14 |
ENSDART00000148452
|
irx2a
|
iroquois homeobox 2a |
| chr13_-_31878263 | 0.13 |
ENSDART00000180411
|
syt14a
|
synaptotagmin XIVa |
| chr1_+_57311901 | 0.13 |
ENSDART00000149397
|
mycbpap
|
mycbp associated protein |
| chr7_+_65586624 | 0.13 |
ENSDART00000184344
|
mical2a
|
microtubule associated monooxygenase, calponin and LIM domain containing 2a |
| chr1_+_58442694 | 0.13 |
ENSDART00000160897
|
zgc:194906
|
zgc:194906 |
| chr1_-_44314 | 0.12 |
ENSDART00000160050
|
tmem39a
|
transmembrane protein 39A |
| chr13_-_31878043 | 0.12 |
ENSDART00000187720
|
syt14a
|
synaptotagmin XIVa |
| chr18_-_3456117 | 0.12 |
ENSDART00000158508
|
myo7aa
|
myosin VIIAa |
| chr23_+_44633858 | 0.12 |
ENSDART00000180728
|
si:ch73-265d7.2
|
si:ch73-265d7.2 |
| chr19_-_10771558 | 0.11 |
ENSDART00000085165
|
slc9a3.2
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3, tandem duplicate 2 |
| chr11_+_25101220 | 0.11 |
ENSDART00000183700
|
ndrg3a
|
ndrg family member 3a |
| chr8_+_10862353 | 0.10 |
ENSDART00000140717
|
brpf3b
|
bromodomain and PHD finger containing, 3b |
| chr1_+_58067815 | 0.10 |
ENSDART00000156678
|
si:ch211-114l13.4
|
si:ch211-114l13.4 |
| chr4_-_2975461 | 0.10 |
ENSDART00000150794
|
plekha5
|
pleckstrin homology domain containing, family A member 5 |
| chr7_+_34296789 | 0.10 |
ENSDART00000052471
ENSDART00000173798 ENSDART00000173778 |
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr22_+_17399124 | 0.09 |
ENSDART00000145769
|
rabgap1l
|
RAB GTPase activating protein 1-like |
| chr7_-_69983948 | 0.09 |
ENSDART00000185827
|
KCNIP4
|
potassium voltage-gated channel interacting protein 4 |
| chr20_-_26936887 | 0.09 |
ENSDART00000160827
|
ftr79
|
finTRIM family, member 79 |
| chr19_-_8798178 | 0.09 |
ENSDART00000188232
|
cers2a
|
ceramide synthase 2a |
| chr2_+_43750899 | 0.08 |
ENSDART00000135836
|
arhgap12a
|
Rho GTPase activating protein 12a |
| chr20_-_26937453 | 0.08 |
ENSDART00000139756
|
ftr97
|
finTRIM family, member 97 |
| chr19_+_3056450 | 0.07 |
ENSDART00000141324
ENSDART00000082353 |
hsf1
|
heat shock transcription factor 1 |
| chr8_+_1433647 | 0.07 |
ENSDART00000160797
|
efna5b
|
ephrin-A5b |
| chr7_-_51793333 | 0.06 |
ENSDART00000180654
|
BX957362.5
|
|
| chr25_-_35960229 | 0.06 |
ENSDART00000073434
|
snx20
|
sorting nexin 20 |
| chr20_+_53278746 | 0.06 |
ENSDART00000074249
|
gpr6
|
G protein-coupled receptor 6 |
| chr1_-_43782 | 0.06 |
ENSDART00000168660
|
tmem39a
|
transmembrane protein 39A |
| chr1_+_58303892 | 0.06 |
ENSDART00000147678
|
CR769768.1
|
|
| chr9_+_38502524 | 0.05 |
ENSDART00000087229
|
lmln
|
leishmanolysin-like (metallopeptidase M8 family) |
| chr20_+_15015557 | 0.05 |
ENSDART00000039345
|
myoc
|
myocilin |
| chr7_+_13988075 | 0.05 |
ENSDART00000186812
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr5_+_29851433 | 0.04 |
ENSDART00000143434
|
ubash3ba
|
ubiquitin associated and SH3 domain containing Ba |
| chr20_+_19793620 | 0.04 |
ENSDART00000136317
|
chrna2b
|
cholinergic receptor, nicotinic, alpha 2b (neuronal) |
| chr3_+_1179601 | 0.04 |
ENSDART00000173378
|
triobpb
|
TRIO and F-actin binding protein b |
| chr6_+_40523370 | 0.04 |
ENSDART00000033819
|
prkcda
|
protein kinase C, delta a |
| chr8_+_37111643 | 0.03 |
ENSDART00000061336
|
ren
|
renin |
| chr25_-_27665978 | 0.03 |
ENSDART00000123590
|
si:ch211-91p5.3
|
si:ch211-91p5.3 |
| chr12_-_22509069 | 0.03 |
ENSDART00000179755
ENSDART00000109707 |
neurl4
|
neuralized E3 ubiquitin protein ligase 4 |
| chr9_-_32912638 | 0.03 |
ENSDART00000110582
|
fam160a2
|
family with sequence similarity 160, member A2 |
| chr2_-_13382726 | 0.03 |
ENSDART00000017787
|
nmur1b
|
neuromedin U receptor 1b |
| chr23_-_45319592 | 0.02 |
ENSDART00000189861
|
ccdc171
|
coiled-coil domain containing 171 |
| chr7_-_2090594 | 0.02 |
ENSDART00000183166
|
si:cabz01007802.1
|
si:cabz01007802.1 |
| chr6_+_8176486 | 0.02 |
ENSDART00000193308
|
nfil3-5
|
nuclear factor, interleukin 3 regulated, member 5 |
| chr5_+_19479200 | 0.02 |
ENSDART00000137703
|
aldh3b2
|
aldehyde dehydrogenase 3 family, member B2 |
| chr3_-_18805225 | 0.02 |
ENSDART00000133471
ENSDART00000131758 |
msrb1a
|
methionine sulfoxide reductase B1a |
| chr7_-_46744080 | 0.02 |
ENSDART00000193328
|
tshz3b
|
teashirt zinc finger homeobox 3b |
| chr25_+_22017182 | 0.02 |
ENSDART00000156517
|
si:dkey-217l24.1
|
si:dkey-217l24.1 |
| chr18_+_43183749 | 0.02 |
ENSDART00000151166
|
nectin1b
|
nectin cell adhesion molecule 1b |
| chr11_+_24314148 | 0.01 |
ENSDART00000171491
|
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr1_+_58119117 | 0.01 |
ENSDART00000146788
|
si:ch211-15j1.5
|
si:ch211-15j1.5 |
| chr19_+_19652439 | 0.01 |
ENSDART00000165934
|
hibadha
|
3-hydroxyisobutyrate dehydrogenase a |
| chr1_+_58260886 | 0.00 |
ENSDART00000110453
|
si:dkey-222h21.8
|
si:dkey-222h21.8 |
| chr19_+_7595924 | 0.00 |
ENSDART00000140083
|
s100s
|
S100 calcium binding protein S |
| chr19_-_27564980 | 0.00 |
ENSDART00000171967
|
si:dkeyp-46h3.8
|
si:dkeyp-46h3.8 |
| chr25_+_12012 | 0.00 |
ENSDART00000170348
|
cntn1a
|
contactin 1a |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr2c1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0009750 | response to fructose(GO:0009750) |
| 0.5 | 1.4 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.2 | 1.2 | GO:1901073 | chitin biosynthetic process(GO:0006031) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.2 | 1.1 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.2 | 1.1 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.2 | 0.6 | GO:0051182 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.1 | 0.6 | GO:0006837 | serotonin transport(GO:0006837) serotonin uptake(GO:0051610) |
| 0.1 | 0.8 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.1 | 0.7 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 0.1 | 0.5 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 1.2 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.1 | 0.3 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.3 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.0 | 0.4 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.3 | GO:0001765 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.9 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.2 | GO:0072584 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.2 | GO:1902902 | negative regulation of autophagosome maturation(GO:1901097) negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.3 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.7 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.5 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.4 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.5 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 2.0 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.1 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.3 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.4 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.0 | 0.5 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.2 | 1.4 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.2 | 1.2 | GO:0030428 | cell septum(GO:0030428) |
| 0.1 | 1.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 0.5 | GO:0071914 | prominosome(GO:0071914) |
| 0.1 | 0.8 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.4 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.1 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.4 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.7 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.4 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.1 | GO:0031526 | brush border membrane(GO:0031526) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0015222 | serotonin:sodium symporter activity(GO:0005335) serotonin transmembrane transporter activity(GO:0015222) |
| 0.2 | 1.1 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.2 | 0.5 | GO:0071077 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.2 | 0.6 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.2 | 0.5 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.2 | 1.2 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.1 | 0.8 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 0.5 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 1.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.5 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.1 | 0.7 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.8 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.1 | 0.8 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 1.9 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.5 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 2.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.3 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.4 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.4 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.4 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.0 | 2.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 4.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.5 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.3 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.3 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.7 | GO:0044325 | ion channel binding(GO:0044325) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.0 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 1.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.1 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |