Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
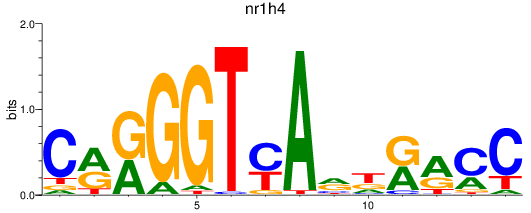
Results for nr1h4
Z-value: 0.30
Transcription factors associated with nr1h4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr1h4
|
ENSDARG00000057741 | nuclear receptor subfamily 1, group H, member 4 |
|
nr1h4
|
ENSDARG00000113600 | nuclear receptor subfamily 1, group H, member 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr1h4 | dr11_v1_chr18_+_15644559_15644559 | -0.39 | 1.0e-01 | Click! |
Activity profile of nr1h4 motif
Sorted Z-values of nr1h4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_15890558 | 0.45 |
ENSDART00000144032
|
si:dkey-14o1.20
|
si:dkey-14o1.20 |
| chr5_+_20693724 | 0.42 |
ENSDART00000141368
|
si:ch211-240b21.2
|
si:ch211-240b21.2 |
| chr5_+_28260158 | 0.37 |
ENSDART00000181434
|
ncaph
|
non-SMC condensin I complex, subunit H |
| chr11_-_22372072 | 0.31 |
ENSDART00000065996
|
tmem183a
|
transmembrane protein 183A |
| chr9_-_1986014 | 0.30 |
ENSDART00000142842
|
hoxd12a
|
homeobox D12a |
| chr1_-_39859626 | 0.29 |
ENSDART00000053763
|
dctd
|
dCMP deaminase |
| chr21_-_19918286 | 0.28 |
ENSDART00000180816
|
ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
| chr1_+_39859782 | 0.28 |
ENSDART00000149984
|
irf2a
|
interferon regulatory factor 2a |
| chr7_-_59159253 | 0.26 |
ENSDART00000159285
|
haus6
|
HAUS augmin-like complex, subunit 6 |
| chr11_-_14924138 | 0.25 |
ENSDART00000168348
|
si:dkey-6d5.1
|
si:dkey-6d5.1 |
| chr16_-_7228276 | 0.25 |
ENSDART00000149030
|
nt5c3a
|
5'-nucleotidase, cytosolic IIIA |
| chr2_+_371759 | 0.23 |
ENSDART00000153788
|
si:dkey-33c14.7
|
si:dkey-33c14.7 |
| chr1_-_17650223 | 0.23 |
ENSDART00000043484
|
si:dkey-256e7.5
|
si:dkey-256e7.5 |
| chr5_-_69571572 | 0.22 |
ENSDART00000145966
|
mapkapk5
|
mitogen-activated protein kinase-activated protein kinase 5 |
| chr10_+_28428222 | 0.21 |
ENSDART00000135003
|
si:ch211-222e20.4
|
si:ch211-222e20.4 |
| chr9_-_27717006 | 0.20 |
ENSDART00000146860
|
gtf2e1
|
general transcription factor IIE, polypeptide 1, alpha |
| chr19_-_18855513 | 0.20 |
ENSDART00000162708
|
ppt2
|
palmitoyl-protein thioesterase 2 |
| chr9_+_38158570 | 0.19 |
ENSDART00000059549
ENSDART00000133060 |
nifk
|
nucleolar protein interacting with the FHA domain of MKI67 |
| chr10_-_21542702 | 0.19 |
ENSDART00000146761
ENSDART00000134502 |
zgc:165539
|
zgc:165539 |
| chr2_+_10160484 | 0.19 |
ENSDART00000136018
|
nsun4
|
NOL1/NOP2/Sun domain family, member 4 |
| chr18_+_30441740 | 0.18 |
ENSDART00000189074
|
gse1
|
Gse1 coiled-coil protein |
| chr6_+_27418541 | 0.18 |
ENSDART00000187410
|
crocc2
|
ciliary rootlet coiled-coil, rootletin family member 2 |
| chr7_+_22792132 | 0.18 |
ENSDART00000135207
ENSDART00000146801 |
rbm4.3
|
RNA binding motif protein 4.3 |
| chr15_+_2857556 | 0.18 |
ENSDART00000157758
|
mre11a
|
MRE11 homolog A, double strand break repair nuclease |
| chr16_+_53203370 | 0.17 |
ENSDART00000154669
|
si:ch211-269k10.2
|
si:ch211-269k10.2 |
| chr4_-_16545085 | 0.17 |
ENSDART00000033188
|
btg1
|
B-cell translocation gene 1, anti-proliferative |
| chr8_-_19342111 | 0.17 |
ENSDART00000138881
|
rgl1
|
ral guanine nucleotide dissociation stimulator-like 1 |
| chr7_+_52811291 | 0.17 |
ENSDART00000192900
|
CT030160.1
|
|
| chr24_+_28448387 | 0.17 |
ENSDART00000154077
ENSDART00000018037 |
arhgap29a
|
Rho GTPase activating protein 29a |
| chr13_-_30713236 | 0.17 |
ENSDART00000112372
ENSDART00000142221 |
tmem72
|
transmembrane protein 72 |
| chr19_-_32493866 | 0.16 |
ENSDART00000052090
|
fuca1.2
|
alpha-L-fucosidase 1, tandem duplicate 2 |
| chr25_-_17503393 | 0.16 |
ENSDART00000061726
|
trim35-40
|
tripartite motif containing 35-40 |
| chr2_+_37838259 | 0.16 |
ENSDART00000136796
|
parp2
|
poly (ADP-ribose) polymerase 2 |
| chr21_+_261490 | 0.16 |
ENSDART00000177919
|
jak2a
|
Janus kinase 2a |
| chr21_+_12056934 | 0.16 |
ENSDART00000125380
|
zgc:162344
|
zgc:162344 |
| chr10_-_41397663 | 0.16 |
ENSDART00000158038
|
gpat4
|
glycerol-3-phosphate acyltransferase 4 |
| chr7_+_34688527 | 0.16 |
ENSDART00000108473
|
plekhg4
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4 |
| chr13_+_2894536 | 0.16 |
ENSDART00000183678
|
CABZ01087629.1
|
|
| chr22_+_8313513 | 0.15 |
ENSDART00000181169
ENSDART00000103911 |
CABZ01077217.1
|
|
| chr19_-_18855717 | 0.15 |
ENSDART00000158192
|
ppt2
|
palmitoyl-protein thioesterase 2 |
| chr5_+_35786141 | 0.15 |
ENSDART00000022043
ENSDART00000127383 |
stard8
|
StAR-related lipid transfer (START) domain containing 8 |
| chr7_+_39399747 | 0.14 |
ENSDART00000147037
|
tnni2b.1
|
troponin I type 2b (skeletal, fast), tandem duplicate 1 |
| chr3_-_52661242 | 0.14 |
ENSDART00000138018
|
zgc:113210
|
zgc:113210 |
| chr12_-_18414536 | 0.14 |
ENSDART00000132956
ENSDART00000078724 |
atpaf2
|
ATP synthase mitochondrial F1 complex assembly factor 2 |
| chr18_+_46151505 | 0.14 |
ENSDART00000015034
ENSDART00000141287 |
blvrb
|
biliverdin reductase B |
| chr14_-_33308138 | 0.14 |
ENSDART00000136442
ENSDART00000139615 |
sept6
|
septin 6 |
| chr15_+_20530649 | 0.14 |
ENSDART00000186312
|
tnfaip1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr16_-_4769877 | 0.14 |
ENSDART00000149421
ENSDART00000054078 |
rpa2
|
replication protein A2 |
| chr21_+_27370671 | 0.14 |
ENSDART00000009234
ENSDART00000142071 |
rbm14a
|
RNA binding motif protein 14a |
| chr10_+_3153973 | 0.14 |
ENSDART00000183223
|
hic2
|
hypermethylated in cancer 2 |
| chr1_-_9130136 | 0.13 |
ENSDART00000113013
|
palb2
|
partner and localizer of BRCA2 |
| chr23_+_28809002 | 0.13 |
ENSDART00000134121
ENSDART00000183661 |
pex14
|
peroxisomal biogenesis factor 14 |
| chr15_-_15910226 | 0.13 |
ENSDART00000154219
|
synrg
|
synergin, gamma |
| chr5_-_38777852 | 0.13 |
ENSDART00000131603
|
si:dkey-58f10.4
|
si:dkey-58f10.4 |
| chr24_-_36337395 | 0.13 |
ENSDART00000154402
|
si:ch211-40k21.9
|
si:ch211-40k21.9 |
| chr5_-_69716501 | 0.13 |
ENSDART00000158956
|
mob1a
|
MOB kinase activator 1A |
| chr12_+_34119439 | 0.13 |
ENSDART00000032821
|
cyth1b
|
cytohesin 1b |
| chr3_-_19899914 | 0.12 |
ENSDART00000134969
|
rnd2
|
Rho family GTPase 2 |
| chr1_+_44941031 | 0.12 |
ENSDART00000141145
|
si:dkey-9i23.16
|
si:dkey-9i23.16 |
| chr11_+_24340166 | 0.12 |
ENSDART00000188719
|
rbm39a
|
RNA binding motif protein 39a |
| chr1_+_6646529 | 0.12 |
ENSDART00000144641
ENSDART00000103701 ENSDART00000138919 |
ube2f
|
ubiquitin-conjugating enzyme E2F (putative) |
| chr4_-_18455480 | 0.12 |
ENSDART00000150221
|
sco2
|
SCO2 cytochrome c oxidase assembly protein |
| chr2_+_314249 | 0.12 |
ENSDART00000082086
|
zgc:113452
|
zgc:113452 |
| chr21_-_5879897 | 0.12 |
ENSDART00000184034
|
rpl35
|
ribosomal protein L35 |
| chr8_+_10305400 | 0.12 |
ENSDART00000172400
|
pim1
|
Pim-1 proto-oncogene, serine/threonine kinase |
| chr14_+_34514336 | 0.12 |
ENSDART00000024440
|
foxi3b
|
forkhead box I3b |
| chr18_+_34181655 | 0.12 |
ENSDART00000130831
ENSDART00000109535 |
gmps
|
guanine monophosphate synthase |
| chr8_+_7315625 | 0.12 |
ENSDART00000135655
ENSDART00000181048 |
selenoh
|
selenoprotein H |
| chr6_+_9677420 | 0.11 |
ENSDART00000064995
|
sumo1
|
small ubiquitin-like modifier 1 |
| chr5_+_69716458 | 0.11 |
ENSDART00000159594
|
mthfd2
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase |
| chr10_+_44692272 | 0.10 |
ENSDART00000157458
|
ubc
|
ubiquitin C |
| chr9_-_25255490 | 0.10 |
ENSDART00000141502
|
htr2aa
|
5-hydroxytryptamine (serotonin) receptor 2A, genome duplicate a |
| chr23_+_43954809 | 0.10 |
ENSDART00000164080
|
corin
|
corin, serine peptidase |
| chr3_+_24595922 | 0.10 |
ENSDART00000169405
|
si:dkey-68o6.5
|
si:dkey-68o6.5 |
| chr6_+_11680726 | 0.10 |
ENSDART00000186732
|
calcrlb
|
calcitonin receptor-like b |
| chr19_+_2685779 | 0.10 |
ENSDART00000160533
ENSDART00000097531 |
tomm7
|
translocase of outer mitochondrial membrane 7 homolog (yeast) |
| chr2_-_37281748 | 0.10 |
ENSDART00000137598
|
nadkb
|
NAD kinase b |
| chr16_+_5901835 | 0.10 |
ENSDART00000060519
|
ulk4
|
unc-51 like kinase 4 |
| chr11_-_18017287 | 0.10 |
ENSDART00000155443
|
qrich1
|
glutamine-rich 1 |
| chr10_-_27197044 | 0.10 |
ENSDART00000137928
|
auts2a
|
autism susceptibility candidate 2a |
| chr11_+_5842632 | 0.10 |
ENSDART00000111374
ENSDART00000158599 |
ndufs7
|
NADH dehydrogenase (ubiquinone) Fe-S protein 7, (NADH-coenzyme Q reductase) |
| chr17_-_36529016 | 0.10 |
ENSDART00000025019
|
colec11
|
collectin sub-family member 11 |
| chr17_-_36529449 | 0.10 |
ENSDART00000187252
|
colec11
|
collectin sub-family member 11 |
| chr19_-_20199167 | 0.10 |
ENSDART00000155527
|
si:ch211-155k24.9
|
si:ch211-155k24.9 |
| chr3_+_24062748 | 0.09 |
ENSDART00000188574
|
cbx1a
|
chromobox homolog 1a (HP1 beta homolog Drosophila) |
| chr7_-_41338923 | 0.09 |
ENSDART00000099138
|
ncf2
|
neutrophil cytosolic factor 2 |
| chr21_+_12036238 | 0.09 |
ENSDART00000102463
ENSDART00000155426 |
zgc:162344
|
zgc:162344 |
| chr6_-_40312091 | 0.09 |
ENSDART00000154878
|
col7a1
|
collagen, type VII, alpha 1 |
| chr7_-_32980017 | 0.09 |
ENSDART00000113744
|
pkp3b
|
plakophilin 3b |
| chr1_-_11689525 | 0.09 |
ENSDART00000090689
|
brat1
|
BRCA1-associated ATM activator 1 |
| chr15_+_24563504 | 0.09 |
ENSDART00000140658
ENSDART00000130589 ENSDART00000045549 |
dhrs13b
|
dehydrogenase/reductase (SDR family) member 13b |
| chr11_-_6974022 | 0.09 |
ENSDART00000172851
|
COMP
|
si:ch211-43f4.1 |
| chr8_+_26879358 | 0.09 |
ENSDART00000132485
|
rimkla
|
ribosomal modification protein rimK-like family member A |
| chr24_+_28528000 | 0.08 |
ENSDART00000155924
|
arhgap29a
|
Rho GTPase activating protein 29a |
| chr6_+_15145123 | 0.08 |
ENSDART00000063648
|
nck2b
|
NCK adaptor protein 2b |
| chr10_+_20070178 | 0.08 |
ENSDART00000027612
ENSDART00000145264 ENSDART00000172713 |
xpo7
|
exportin 7 |
| chr20_-_28352352 | 0.08 |
ENSDART00000128806
|
ino80
|
INO80 complex subunit |
| chr10_+_44363195 | 0.08 |
ENSDART00000161744
|
kmt5aa
|
lysine methyltransferase 5Aa |
| chr11_+_691734 | 0.08 |
ENSDART00000191463
|
timp4.1
|
TIMP metallopeptidase inhibitor 4, tandem duplicate 1 |
| chr21_-_32082130 | 0.08 |
ENSDART00000003978
ENSDART00000182050 |
mat2b
|
methionine adenosyltransferase II, beta |
| chr9_-_48184823 | 0.08 |
ENSDART00000180264
|
klhl23
|
kelch-like family member 23 |
| chr23_+_20431388 | 0.08 |
ENSDART00000132920
ENSDART00000102963 ENSDART00000109899 ENSDART00000140219 |
slc35c2
|
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr7_+_37359004 | 0.08 |
ENSDART00000192134
ENSDART00000189239 |
sall1a
|
spalt-like transcription factor 1a |
| chr7_-_52531252 | 0.08 |
ENSDART00000174369
|
tcf12
|
transcription factor 12 |
| chr13_-_7766758 | 0.07 |
ENSDART00000171831
|
h2afy2
|
H2A histone family, member Y2 |
| chr17_+_15111319 | 0.07 |
ENSDART00000185099
|
CU464134.2
|
|
| chr5_-_25072607 | 0.07 |
ENSDART00000145061
|
pnpla7b
|
patatin-like phospholipase domain containing 7b |
| chr12_-_25201576 | 0.07 |
ENSDART00000077188
|
cox7a3
|
cytochrome c oxidase subunit VIIa polypeptide 3 |
| chr16_-_20312146 | 0.07 |
ENSDART00000134980
|
si:dkeyp-86h10.3
|
si:dkeyp-86h10.3 |
| chr5_+_65946222 | 0.07 |
ENSDART00000190969
ENSDART00000161578 |
mymk
|
myomaker, myoblast fusion factor |
| chr6_+_38626684 | 0.07 |
ENSDART00000086533
|
atp10a
|
ATPase phospholipid transporting 10A |
| chr15_-_18091157 | 0.07 |
ENSDART00000153848
|
phldb1b
|
pleckstrin homology-like domain, family B, member 1b |
| chr21_-_41620419 | 0.07 |
ENSDART00000180642
|
pcyox1l
|
prenylcysteine oxidase 1 like |
| chr2_-_37862380 | 0.07 |
ENSDART00000186005
|
si:ch211-284o19.8
|
si:ch211-284o19.8 |
| chr21_+_21263988 | 0.07 |
ENSDART00000089651
ENSDART00000108978 |
ccdc61
|
coiled-coil domain containing 61 |
| chr5_+_38886499 | 0.07 |
ENSDART00000076845
|
fras1
|
Fraser extracellular matrix complex subunit 1 |
| chr20_-_19864131 | 0.07 |
ENSDART00000057819
|
ptk2bb
|
protein tyrosine kinase 2 beta, b |
| chr8_-_48675411 | 0.06 |
ENSDART00000165081
|
pimr183
|
Pim proto-oncogene, serine/threonine kinase, related 183 |
| chr25_+_30196039 | 0.06 |
ENSDART00000005299
|
hsd17b12a
|
hydroxysteroid (17-beta) dehydrogenase 12a |
| chr25_-_24046870 | 0.06 |
ENSDART00000047569
|
igf2b
|
insulin-like growth factor 2b |
| chr16_+_14684916 | 0.06 |
ENSDART00000138611
|
col14a1a
|
collagen, type XIV, alpha 1a |
| chr6_-_1514767 | 0.06 |
ENSDART00000067586
|
chchd6b
|
coiled-coil-helix-coiled-coil-helix domain containing 6b |
| chr17_-_7028418 | 0.06 |
ENSDART00000188305
ENSDART00000187895 |
sash1b
|
SAM and SH3 domain containing 1b |
| chr15_-_19705707 | 0.06 |
ENSDART00000047643
|
sytl2b
|
synaptotagmin-like 2b |
| chr7_+_53755054 | 0.06 |
ENSDART00000181629
|
neo1a
|
neogenin 1a |
| chr8_-_13013123 | 0.06 |
ENSDART00000147802
|
dennd2da
|
DENN/MADD domain containing 2Da |
| chr5_+_54400971 | 0.06 |
ENSDART00000169695
|
bspry
|
B-box and SPRY domain containing |
| chr17_-_50018133 | 0.06 |
ENSDART00000112267
|
filip1a
|
filamin A interacting protein 1a |
| chr7_+_24523017 | 0.06 |
ENSDART00000077047
|
btr09
|
bloodthirsty-related gene family, member 9 |
| chr21_+_13383413 | 0.06 |
ENSDART00000151345
|
zgc:113162
|
zgc:113162 |
| chr1_-_21538329 | 0.06 |
ENSDART00000182798
ENSDART00000193518 ENSDART00000183873 ENSDART00000183376 |
wdr19
|
WD repeat domain 19 |
| chr9_+_48007081 | 0.06 |
ENSDART00000060593
ENSDART00000099835 |
zgc:92380
|
zgc:92380 |
| chr5_+_31350927 | 0.06 |
ENSDART00000028237
|
camkk1a
|
calcium/calmodulin-dependent protein kinase kinase 1, alpha a |
| chr12_-_14143344 | 0.06 |
ENSDART00000152742
|
buc2l
|
bucky ball 2-like |
| chr4_-_77114795 | 0.06 |
ENSDART00000144849
|
CU467646.2
|
|
| chr3_-_34027178 | 0.05 |
ENSDART00000170201
ENSDART00000151408 |
ighv1-4
ighv14-1
|
immunoglobulin heavy variable 1-4 immunoglobulin heavy variable 14-1 |
| chr21_+_29179887 | 0.05 |
ENSDART00000161941
|
si:ch211-57b15.1
|
si:ch211-57b15.1 |
| chr9_-_9705512 | 0.05 |
ENSDART00000149021
|
gpr156
|
G protein-coupled receptor 156 |
| chr22_-_876506 | 0.05 |
ENSDART00000137522
|
cept1b
|
choline/ethanolamine phosphotransferase 1b |
| chr15_-_31000275 | 0.05 |
ENSDART00000132493
ENSDART00000100183 |
lgals9l6
lgals9l5
|
lectin, galactoside-binding, soluble, 9 (galectin 9)-like 6 lectin, galactoside-binding, soluble, 9 (galectin 9)-like 5 |
| chr7_-_40630698 | 0.05 |
ENSDART00000134547
|
ube3c
|
ubiquitin protein ligase E3C |
| chr22_+_10645088 | 0.05 |
ENSDART00000142265
|
rassf1
|
Ras association (RalGDS/AF-6) domain family 1 |
| chr21_-_22664554 | 0.05 |
ENSDART00000161438
|
gig2k
|
grass carp reovirus (GCRV)-induced gene 2k |
| chr19_-_23369282 | 0.05 |
ENSDART00000147824
|
pimr90
|
Pim proto-oncogene, serine/threonine kinase, related 90 |
| chr5_+_50913034 | 0.05 |
ENSDART00000149787
|
col4a3bpa
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein a |
| chr2_-_27651674 | 0.05 |
ENSDART00000177402
|
tgs1
|
trimethylguanosine synthase 1 |
| chr19_+_32166702 | 0.05 |
ENSDART00000021798
|
fabp11a
|
fatty acid binding protein 11a |
| chr23_+_22597624 | 0.05 |
ENSDART00000054337
|
gpr157
|
G protein-coupled receptor 157 |
| chr12_+_695619 | 0.05 |
ENSDART00000161691
|
abcc3
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
| chr4_-_5597167 | 0.05 |
ENSDART00000132431
|
vegfab
|
vascular endothelial growth factor Ab |
| chr21_+_44112914 | 0.04 |
ENSDART00000062836
|
fgf1b
|
fibroblast growth factor 1b |
| chr25_+_23591990 | 0.04 |
ENSDART00000151938
ENSDART00000089199 |
cpt1ab
|
carnitine palmitoyltransferase 1Ab (liver) |
| chr23_-_22598002 | 0.04 |
ENSDART00000146966
|
si:ch211-28e16.5
|
si:ch211-28e16.5 |
| chr24_+_25467465 | 0.04 |
ENSDART00000189933
|
smpx
|
small muscle protein, X-linked |
| chr9_-_28616436 | 0.04 |
ENSDART00000136985
|
si:ch73-7i4.2
|
si:ch73-7i4.2 |
| chr17_+_30843881 | 0.04 |
ENSDART00000149600
ENSDART00000148547 |
tpp1
|
tripeptidyl peptidase I |
| chr23_-_44723102 | 0.04 |
ENSDART00000129138
|
mogat3a
|
monoacylglycerol O-acyltransferase 3a |
| chr25_+_12494079 | 0.04 |
ENSDART00000163508
|
map1lc3b
|
microtubule-associated protein 1 light chain 3 beta |
| chr19_+_30990815 | 0.04 |
ENSDART00000134645
|
sync
|
syncoilin, intermediate filament protein |
| chr6_+_52869892 | 0.04 |
ENSDART00000146143
|
si:dkeyp-3f10.16
|
si:dkeyp-3f10.16 |
| chr20_+_7243879 | 0.04 |
ENSDART00000122298
|
bsnd
|
barttin CLCNK-type chloride channel accessory beta subunit |
| chr11_-_41786522 | 0.04 |
ENSDART00000191066
|
CR847970.1
|
|
| chr22_-_36926342 | 0.04 |
ENSDART00000151804
|
si:dkey-37m8.11
|
si:dkey-37m8.11 |
| chr7_-_24838367 | 0.04 |
ENSDART00000139455
ENSDART00000012483 ENSDART00000131530 |
fam113
|
family with sequence similarity 113 |
| chr5_-_19833310 | 0.04 |
ENSDART00000138186
|
trpv4
|
transient receptor potential cation channel, subfamily V, member 4 |
| chr9_-_31090593 | 0.04 |
ENSDART00000089136
|
gpr18
|
G protein-coupled receptor 18 |
| chr19_-_1929310 | 0.04 |
ENSDART00000144113
|
si:ch211-149a19.3
|
si:ch211-149a19.3 |
| chr7_+_38667066 | 0.03 |
ENSDART00000013394
|
mtch2
|
mitochondrial carrier homolog 2 |
| chr8_+_53311965 | 0.03 |
ENSDART00000130104
|
gnb1a
|
guanine nucleotide binding protein (G protein), beta polypeptide 1a |
| chr22_+_9294336 | 0.03 |
ENSDART00000193694
|
si:ch211-250k18.7
|
si:ch211-250k18.7 |
| chr13_-_23665580 | 0.03 |
ENSDART00000144282
|
map3k21
|
mitogen-activated protein kinase kinase kinase 21 |
| chr11_-_544187 | 0.03 |
ENSDART00000173161
|
CU695207.1
|
|
| chr22_+_24318131 | 0.03 |
ENSDART00000187360
ENSDART00000165618 |
ccdc50
|
coiled-coil domain containing 50 |
| chr15_-_2857961 | 0.03 |
ENSDART00000033263
|
ankrd49
|
ankyrin repeat domain 49 |
| chr20_-_4738101 | 0.03 |
ENSDART00000050201
ENSDART00000152559 ENSDART00000053858 ENSDART00000125620 |
papola
|
poly(A) polymerase alpha |
| chr10_-_26202766 | 0.03 |
ENSDART00000136393
|
fhdc3
|
FH2 domain containing 3 |
| chr8_-_28339151 | 0.03 |
ENSDART00000148765
ENSDART00000149173 ENSDART00000150113 |
kdm5bb
|
lysine (K)-specific demethylase 5Bb |
| chr23_+_27779452 | 0.03 |
ENSDART00000134785
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr5_+_1219187 | 0.03 |
ENSDART00000129490
|
gb:bc139872
|
expressed sequence BC139872 |
| chr20_+_46311707 | 0.03 |
ENSDART00000184743
|
flvcr2b
|
feline leukemia virus subgroup C cellular receptor family, member 2b |
| chr22_+_25590391 | 0.03 |
ENSDART00000178133
|
aars2
|
alanyl-tRNA synthetase 2, mitochondrial (putative) |
| chr22_+_34663328 | 0.03 |
ENSDART00000183912
|
si:ch73-304f21.1
|
si:ch73-304f21.1 |
| chr17_+_16192486 | 0.02 |
ENSDART00000156832
|
kif13ba
|
kinesin family member 13Ba |
| chr7_-_1918971 | 0.02 |
ENSDART00000181759
|
CABZ01040626.1
|
|
| chr5_-_25665254 | 0.02 |
ENSDART00000131515
|
tmc1
|
transmembrane channel-like 1 |
| chr23_+_7471072 | 0.02 |
ENSDART00000135551
|
si:ch211-200e2.1
|
si:ch211-200e2.1 |
| chr1_+_58353661 | 0.02 |
ENSDART00000140074
|
si:dkey-222h21.2
|
si:dkey-222h21.2 |
| chr13_+_36958086 | 0.02 |
ENSDART00000024386
|
frmd6
|
FERM domain containing 6 |
| chr25_+_245018 | 0.02 |
ENSDART00000155344
|
zgc:92481
|
zgc:92481 |
| chr23_+_35684756 | 0.02 |
ENSDART00000132320
|
pmelb
|
premelanosome protein b |
| chr7_+_29153018 | 0.02 |
ENSDART00000138065
|
tmppe
|
transmembrane protein with metallophosphoesterase domain |
| chr3_-_43662495 | 0.02 |
ENSDART00000166729
|
axin1
|
axin 1 |
| chr18_+_31056645 | 0.02 |
ENSDART00000159316
|
mvda
|
mevalonate (diphospho) decarboxylase a |
| chr4_-_38281909 | 0.02 |
ENSDART00000162448
|
si:ch211-229l10.9
|
si:ch211-229l10.9 |
| chr4_+_1530287 | 0.02 |
ENSDART00000067446
|
slc38a4
|
solute carrier family 38, member 4 |
| chr15_+_6652396 | 0.02 |
ENSDART00000192813
ENSDART00000157678 |
nop53
|
NOP53 ribosome biogenesis factor |
| chr15_-_17619306 | 0.02 |
ENSDART00000184011
|
adamts15b
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15b |
| chr1_+_58139102 | 0.02 |
ENSDART00000134826
|
si:ch211-15j1.4
|
si:ch211-15j1.4 |
| chr10_-_36207737 | 0.02 |
ENSDART00000159366
|
or109-13
|
odorant receptor, family D, subfamily 109, member 13 |
| chr14_-_8787525 | 0.02 |
ENSDART00000160848
|
tgfb2l
|
transforming growth factor, beta 2, like |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr1h4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.1 | 0.3 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 0.2 | GO:0001120 | DNA-templated transcriptional open complex formation(GO:0001112) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) protein-DNA complex remodeling(GO:0001120) |
| 0.0 | 0.1 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 0.1 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.0 | 0.4 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.1 | GO:0071276 | cellular response to cadmium ion(GO:0071276) |
| 0.0 | 0.2 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.1 | GO:0015990 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) electron transport coupled proton transport(GO:0015990) |
| 0.0 | 0.1 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.2 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.1 | GO:0014743 | regulation of muscle hypertrophy(GO:0014743) |
| 0.0 | 0.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.1 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.0 | 0.1 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.2 | GO:0019730 | antimicrobial humoral response(GO:0019730) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:0030327 | prenylated protein catabolic process(GO:0030327) prenylcysteine catabolic process(GO:0030328) prenylcysteine metabolic process(GO:0030329) |
| 0.0 | 0.2 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.1 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.0 | 0.0 | GO:0071470 | cellular response to osmotic stress(GO:0071470) cellular response to fluid shear stress(GO:0071498) |
| 0.0 | 0.2 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.1 | GO:2000562 | negative regulation of interferon-gamma production(GO:0032689) CD4-positive, alpha-beta T cell proliferation(GO:0035739) regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1990498 | mitotic spindle microtubule(GO:1990498) |
| 0.0 | 0.4 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.1 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.0 | 0.2 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0042806 | fucose binding(GO:0042806) |
| 0.1 | 0.2 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.0 | 0.1 | GO:0001635 | adrenomedullin receptor activity(GO:0001605) calcitonin gene-related peptide receptor activity(GO:0001635) |
| 0.0 | 0.2 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) citrate-L-glutamate ligase activity(GO:0072591) |
| 0.0 | 0.3 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:0001735 | prenylcysteine oxidase activity(GO:0001735) |
| 0.0 | 0.2 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.0 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.2 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID AURORA B PATHWAY | Aurora B signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.3 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.3 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.1 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |