Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
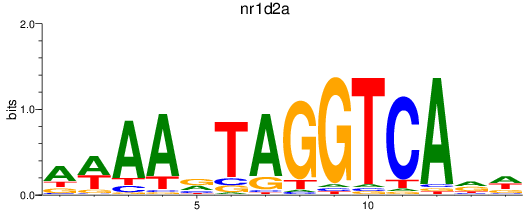
Results for nr1d2a
Z-value: 1.20
Transcription factors associated with nr1d2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr1d2a
|
ENSDARG00000003820 | nuclear receptor subfamily 1, group D, member 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr1d2a | dr11_v1_chr16_+_50089417_50089417 | 0.92 | 3.2e-08 | Click! |
Activity profile of nr1d2a motif
Sorted Z-values of nr1d2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_13244149 | 4.02 |
ENSDART00000186984
ENSDART00000105896 |
atp2a1l
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1, like |
| chr15_+_19682013 | 3.94 |
ENSDART00000127368
|
si:dkey-4p15.5
|
si:dkey-4p15.5 |
| chr5_-_11573490 | 3.62 |
ENSDART00000109577
|
FO704871.1
|
|
| chr17_-_6738538 | 3.48 |
ENSDART00000157125
|
vsnl1b
|
visinin-like 1b |
| chr23_+_19564392 | 3.36 |
ENSDART00000144746
|
atp6ap1lb
|
ATPase H+ transporting accessory protein 1 like b |
| chr2_+_31804582 | 3.12 |
ENSDART00000086646
|
rnf182
|
ring finger protein 182 |
| chr3_+_1016404 | 3.09 |
ENSDART00000189545
|
si:ch1073-464p5.5
|
si:ch1073-464p5.5 |
| chr18_+_907266 | 3.03 |
ENSDART00000171729
|
pkma
|
pyruvate kinase M1/2a |
| chr3_+_35298078 | 3.01 |
ENSDART00000110126
|
cacng3b
|
calcium channel, voltage-dependent, gamma subunit 3b |
| chr25_+_6306885 | 2.91 |
ENSDART00000142705
ENSDART00000067510 |
crabp1a
|
cellular retinoic acid binding protein 1a |
| chr8_-_33114202 | 2.85 |
ENSDART00000098840
|
ralgps1
|
Ral GEF with PH domain and SH3 binding motif 1 |
| chr14_+_17376940 | 2.82 |
ENSDART00000054590
ENSDART00000010148 |
spon2b
|
spondin 2b, extracellular matrix protein |
| chr5_-_67878064 | 2.77 |
ENSDART00000111203
|
tagln3a
|
transgelin 3a |
| chr19_+_4990320 | 2.77 |
ENSDART00000147056
|
zgc:91968
|
zgc:91968 |
| chr7_+_65240227 | 2.66 |
ENSDART00000168287
|
bco1l
|
beta-carotene oxygenase 1, like |
| chr19_+_4990496 | 2.60 |
ENSDART00000151050
ENSDART00000017535 |
zgc:91968
|
zgc:91968 |
| chr13_+_29470442 | 2.59 |
ENSDART00000028417
|
lrit2
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 2 |
| chr15_+_19681718 | 2.48 |
ENSDART00000164803
|
si:dkey-4p15.5
|
si:dkey-4p15.5 |
| chr20_+_16750177 | 2.46 |
ENSDART00000185357
|
calm1b
|
calmodulin 1b |
| chr6_-_39605734 | 2.23 |
ENSDART00000044276
ENSDART00000179059 |
dip2bb
|
disco-interacting protein 2 homolog Bb |
| chr22_-_38480186 | 2.22 |
ENSDART00000171704
|
soul4
|
heme-binding protein soul4 |
| chr12_+_28574863 | 2.22 |
ENSDART00000153284
|
tbkbp1
|
TBK1 binding protein 1 |
| chr5_-_30079434 | 2.15 |
ENSDART00000133981
|
bco2a
|
beta-carotene oxygenase 2a |
| chr20_+_36233873 | 2.15 |
ENSDART00000131867
|
cnih3
|
cornichon family AMPA receptor auxiliary protein 3 |
| chr24_-_32408404 | 2.12 |
ENSDART00000144157
|
si:ch211-56a11.2
|
si:ch211-56a11.2 |
| chr15_-_24178893 | 2.07 |
ENSDART00000077980
|
pipox
|
pipecolic acid oxidase |
| chr25_+_31264155 | 2.04 |
ENSDART00000012256
|
tnni2a.3
|
troponin I type 2a (skeletal, fast), tandem duplicate 3 |
| chr11_+_25430851 | 1.99 |
ENSDART00000164999
ENSDART00000126403 |
si:dkey-13a21.4
|
si:dkey-13a21.4 |
| chr25_+_14165447 | 1.94 |
ENSDART00000145387
|
shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr12_+_1286642 | 1.91 |
ENSDART00000157467
|
pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr16_+_11558868 | 1.85 |
ENSDART00000112497
ENSDART00000180445 |
zgc:198329
|
zgc:198329 |
| chr2_+_33189582 | 1.81 |
ENSDART00000145588
ENSDART00000136330 ENSDART00000139295 ENSDART00000086340 |
rnf220a
|
ring finger protein 220a |
| chr12_-_3756405 | 1.80 |
ENSDART00000150839
|
fam57bb
|
family with sequence similarity 57, member Bb |
| chr1_-_30039331 | 1.77 |
ENSDART00000086935
ENSDART00000143800 |
MARCH4 (1 of many)
|
zgc:153256 |
| chr9_+_13682133 | 1.76 |
ENSDART00000175639
|
mpp4a
|
membrane protein, palmitoylated 4a (MAGUK p55 subfamily member 4) |
| chr12_+_12112384 | 1.75 |
ENSDART00000152431
|
grid1b
|
glutamate receptor, ionotropic, delta 1b |
| chr23_+_44614056 | 1.69 |
ENSDART00000188379
|
eno3
|
enolase 3, (beta, muscle) |
| chr2_-_16380283 | 1.67 |
ENSDART00000149992
|
si:dkey-231j24.3
|
si:dkey-231j24.3 |
| chr21_+_40106448 | 1.58 |
ENSDART00000100166
|
serpinf1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr19_-_8732037 | 1.54 |
ENSDART00000138971
|
si:ch211-39a7.1
|
si:ch211-39a7.1 |
| chr20_-_14680897 | 1.48 |
ENSDART00000063857
ENSDART00000161314 |
scrn2
|
secernin 2 |
| chr21_-_40557281 | 1.47 |
ENSDART00000172327
|
taok1b
|
TAO kinase 1b |
| chr7_+_66048102 | 1.45 |
ENSDART00000104523
|
arntl1b
|
aryl hydrocarbon receptor nuclear translocator-like 1b |
| chr2_+_20430366 | 1.45 |
ENSDART00000155108
|
si:ch211-153l6.6
|
si:ch211-153l6.6 |
| chr21_+_27189490 | 1.43 |
ENSDART00000125349
|
bada
|
BCL2 associated agonist of cell death a |
| chr25_-_27842654 | 1.37 |
ENSDART00000154852
ENSDART00000156906 |
asb15a
|
ankyrin repeat and SOCS box containing 15a |
| chr25_-_1235457 | 1.37 |
ENSDART00000093093
|
coro2bb
|
coronin, actin binding protein, 2Bb |
| chr18_+_22302635 | 1.35 |
ENSDART00000141051
|
carmil2
|
capping protein regulator and myosin 1 linker 2 |
| chr17_+_5793248 | 1.33 |
ENSDART00000153743
|
rp1l1a
|
retinitis pigmentosa 1-like 1a |
| chr11_+_30672282 | 1.33 |
ENSDART00000023981
ENSDART00000188846 |
ttbk1a
|
tau tubulin kinase 1a |
| chr1_+_9708801 | 1.33 |
ENSDART00000189621
|
elfn1b
|
extracellular leucine-rich repeat and fibronectin type III domain containing 1b |
| chr18_-_15559817 | 1.30 |
ENSDART00000061681
|
si:ch211-245j22.3
|
si:ch211-245j22.3 |
| chr7_-_28058442 | 1.28 |
ENSDART00000173842
|
si:ch211-235p24.2
|
si:ch211-235p24.2 |
| chr12_-_32183164 | 1.28 |
ENSDART00000191269
ENSDART00000173549 |
SMIM36
|
si:ch73-256g18.2 |
| chr16_-_24561354 | 1.28 |
ENSDART00000193278
ENSDART00000126274 |
si:ch211-79k12.2
|
si:ch211-79k12.2 |
| chr6_+_57541776 | 1.26 |
ENSDART00000157330
|
necab3
|
N-terminal EF-hand calcium binding protein 3 |
| chr8_-_18899427 | 1.18 |
ENSDART00000079840
|
rorca
|
RAR-related orphan receptor C a |
| chr7_-_2047639 | 1.11 |
ENSDART00000173892
|
si:cabz01007794.1
|
si:cabz01007794.1 |
| chr22_+_21317597 | 1.08 |
ENSDART00000132605
|
shc2
|
SHC (Src homology 2 domain containing) transforming protein 2 |
| chr19_+_2590182 | 1.07 |
ENSDART00000162293
|
si:ch73-345f18.3
|
si:ch73-345f18.3 |
| chr25_+_20188769 | 1.04 |
ENSDART00000142781
|
cald1b
|
caldesmon 1b |
| chr15_+_2559875 | 1.02 |
ENSDART00000178505
|
SH2B2
|
SH2B adaptor protein 2 |
| chr15_+_42599501 | 0.99 |
ENSDART00000177646
|
grik1b
|
glutamate receptor, ionotropic, kainate 1b |
| chr12_-_32013125 | 0.99 |
ENSDART00000153355
|
grin2cb
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2Cb |
| chr9_+_35876927 | 0.99 |
ENSDART00000138834
|
mab21l3
|
mab-21-like 3 |
| chr8_-_3773374 | 0.99 |
ENSDART00000115036
|
bicdl1
|
BICD family like cargo adaptor 1 |
| chr19_-_15335787 | 0.99 |
ENSDART00000187131
|
hivep3a
|
human immunodeficiency virus type I enhancer binding protein 3a |
| chr23_-_44233408 | 0.97 |
ENSDART00000149318
ENSDART00000085528 |
zgc:158659
|
zgc:158659 |
| chr2_+_30721070 | 0.97 |
ENSDART00000099052
|
si:dkey-94e7.2
|
si:dkey-94e7.2 |
| chr2_-_56655769 | 0.95 |
ENSDART00000113589
|
gpx4b
|
glutathione peroxidase 4b |
| chr1_+_55752593 | 0.93 |
ENSDART00000108838
ENSDART00000134770 |
tecrb
|
trans-2,3-enoyl-CoA reductase b |
| chr1_-_44704261 | 0.91 |
ENSDART00000133210
|
si:dkey-28b4.8
|
si:dkey-28b4.8 |
| chr18_+_7594012 | 0.90 |
ENSDART00000062150
|
zgc:77752
|
zgc:77752 |
| chr17_+_20174044 | 0.88 |
ENSDART00000156028
|
si:ch211-248a14.8
|
si:ch211-248a14.8 |
| chr25_-_27843066 | 0.85 |
ENSDART00000179684
ENSDART00000186000 ENSDART00000190065 |
asb15a
|
ankyrin repeat and SOCS box containing 15a |
| chr18_-_39473055 | 0.84 |
ENSDART00000122930
|
scg3
|
secretogranin III |
| chr10_+_2232023 | 0.84 |
ENSDART00000097695
|
cntnap3
|
contactin associated protein like 3 |
| chr22_+_17828267 | 0.84 |
ENSDART00000136016
|
hapln4
|
hyaluronan and proteoglycan link protein 4 |
| chr16_+_30438041 | 0.82 |
ENSDART00000137977
|
pear1
|
platelet endothelial aggregation receptor 1 |
| chr6_+_32415132 | 0.78 |
ENSDART00000155790
|
kank4
|
KN motif and ankyrin repeat domains 4 |
| chr9_-_48937089 | 0.77 |
ENSDART00000193442
|
cers6
|
ceramide synthase 6 |
| chr18_-_35736591 | 0.76 |
ENSDART00000036015
|
ryr1b
|
ryanodine receptor 1b (skeletal) |
| chr18_-_42313798 | 0.75 |
ENSDART00000098639
|
cntn5
|
contactin 5 |
| chr24_+_24809877 | 0.74 |
ENSDART00000185117
|
dnajc5b
|
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
| chr6_+_37752781 | 0.71 |
ENSDART00000154364
|
herc2
|
HECT and RLD domain containing E3 ubiquitin protein ligase 2 |
| chr22_-_29689485 | 0.71 |
ENSDART00000182173
|
pdcd4b
|
programmed cell death 4b |
| chr3_-_2592350 | 0.70 |
ENSDART00000192325
|
si:dkey-217f16.5
|
si:dkey-217f16.5 |
| chr12_+_19305390 | 0.66 |
ENSDART00000183987
ENSDART00000066391 |
csnk1e
|
casein kinase 1, epsilon |
| chr11_-_21303946 | 0.64 |
ENSDART00000185786
|
RASSF5
|
si:dkey-85p17.3 |
| chr17_-_7371564 | 0.58 |
ENSDART00000060336
|
rab32b
|
RAB32b, member RAS oncogene family |
| chr17_+_6625717 | 0.56 |
ENSDART00000190753
ENSDART00000181298 |
BX005321.1
|
|
| chr9_-_8661436 | 0.52 |
ENSDART00000130442
|
col4a1
|
collagen, type IV, alpha 1 |
| chr8_+_42917515 | 0.50 |
ENSDART00000021715
|
slc23a2
|
solute carrier family 23 (ascorbic acid transporter), member 2 |
| chr1_-_21297748 | 0.46 |
ENSDART00000142109
|
npy1r
|
neuropeptide Y receptor Y1 |
| chr21_+_30549512 | 0.45 |
ENSDART00000132831
|
rab38c
|
RAB38c, member of RAS oncogene family |
| chr3_-_2591942 | 0.45 |
ENSDART00000127971
|
si:dkey-217f16.5
|
si:dkey-217f16.5 |
| chr5_-_35888499 | 0.43 |
ENSDART00000193932
|
rxfp2l
|
relaxin/insulin-like family peptide receptor 2, like |
| chr6_-_21726758 | 0.42 |
ENSDART00000083085
|
mtmr14
|
myotubularin related protein 14 |
| chr3_+_60761811 | 0.42 |
ENSDART00000053482
|
tsen54
|
TSEN54 tRNA splicing endonuclease subunit |
| chr9_+_17982737 | 0.41 |
ENSDART00000192569
|
akap11
|
A kinase (PRKA) anchor protein 11 |
| chr8_+_18464235 | 0.40 |
ENSDART00000110571
|
alkal1
|
ALK and LTK ligand 1 |
| chr19_+_30990129 | 0.40 |
ENSDART00000052169
ENSDART00000193376 |
sync
|
syncoilin, intermediate filament protein |
| chr4_-_22519516 | 0.39 |
ENSDART00000130409
ENSDART00000186258 ENSDART00000002851 ENSDART00000123801 |
kdm7aa
|
lysine (K)-specific demethylase 7Aa |
| chr19_+_30990815 | 0.38 |
ENSDART00000134645
|
sync
|
syncoilin, intermediate filament protein |
| chr12_+_7399403 | 0.36 |
ENSDART00000103526
|
bicc1b
|
BicC family RNA binding protein 1b |
| chr5_+_38743644 | 0.35 |
ENSDART00000137658
|
si:dkey-58f10.6
|
si:dkey-58f10.6 |
| chr1_-_49498116 | 0.35 |
ENSDART00000137357
|
zgc:175214
|
zgc:175214 |
| chr18_+_27738349 | 0.35 |
ENSDART00000187816
|
tspan18b
|
tetraspanin 18b |
| chr15_-_34322915 | 0.34 |
ENSDART00000190543
ENSDART00000019651 ENSDART00000193601 |
dgkb
|
diacylglycerol kinase, beta |
| chr2_-_42558549 | 0.34 |
ENSDART00000025997
|
dip2cb
|
disco-interacting protein 2 homolog Cb |
| chr17_-_15528597 | 0.34 |
ENSDART00000150232
|
fyna
|
FYN proto-oncogene, Src family tyrosine kinase a |
| chr11_-_21304452 | 0.34 |
ENSDART00000163008
|
RASSF5
|
si:dkey-85p17.3 |
| chr23_+_41831224 | 0.33 |
ENSDART00000171885
|
scp2b
|
sterol carrier protein 2b |
| chr23_-_5729484 | 0.33 |
ENSDART00000000488
|
tnni1a
|
troponin I type 1a (skeletal, slow) |
| chr3_-_16039619 | 0.32 |
ENSDART00000143324
|
spsb3a
|
splA/ryanodine receptor domain and SOCS box containing 3a |
| chr25_-_17918810 | 0.32 |
ENSDART00000023959
|
arntl1a
|
aryl hydrocarbon receptor nuclear translocator-like 1a |
| chr22_-_6849585 | 0.32 |
ENSDART00000125750
|
FO904898.1
|
|
| chr3_+_57997980 | 0.31 |
ENSDART00000168477
ENSDART00000193840 |
pycr1a
|
pyrroline-5-carboxylate reductase 1a |
| chr12_+_30789611 | 0.31 |
ENSDART00000181501
|
aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr6_-_8244474 | 0.26 |
ENSDART00000151358
|
rgl3a
|
ral guanine nucleotide dissociation stimulator-like 3a |
| chr21_-_2209012 | 0.25 |
ENSDART00000158345
|
zgc:162971
|
zgc:162971 |
| chr5_+_43470544 | 0.25 |
ENSDART00000111587
|
rhobtb2a
|
Rho-related BTB domain containing 2a |
| chr16_+_8716800 | 0.22 |
ENSDART00000124693
ENSDART00000181961 |
cabz01093075.1
|
cabz01093075.1 |
| chr11_+_19159083 | 0.20 |
ENSDART00000138964
|
adamts9
|
ADAM metallopeptidase with thrombospondin type 1 motif, 9 |
| chr23_-_36003282 | 0.19 |
ENSDART00000103150
|
calcoco1a
|
calcium binding and coiled-coil domain 1a |
| chr20_+_44082532 | 0.19 |
ENSDART00000188481
|
BX510352.1
|
|
| chr8_+_14158021 | 0.18 |
ENSDART00000080832
|
si:dkey-6n6.2
|
si:dkey-6n6.2 |
| chr22_-_23000815 | 0.17 |
ENSDART00000137111
|
ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr12_+_9542124 | 0.17 |
ENSDART00000127952
|
p4ha1a
|
prolyl 4-hydroxylase, alpha polypeptide I a |
| chr13_+_30172645 | 0.16 |
ENSDART00000137114
|
pald1b
|
phosphatase domain containing, paladin 1b |
| chr8_+_18463797 | 0.14 |
ENSDART00000148958
|
alkal1
|
ALK and LTK ligand 1 |
| chr8_+_43340995 | 0.13 |
ENSDART00000038566
|
rflna
|
refilin A |
| chr3_+_32698424 | 0.12 |
ENSDART00000055340
|
fus
|
FUS RNA binding protein |
| chr1_+_15204633 | 0.12 |
ENSDART00000188410
|
itln1
|
intelectin 1 |
| chr22_+_36704764 | 0.12 |
ENSDART00000146662
|
trim35-33
|
tripartite motif containing 35-33 |
| chr9_+_28116079 | 0.11 |
ENSDART00000101338
|
idh1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr15_+_17100412 | 0.11 |
ENSDART00000154418
|
relb
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr8_+_21146262 | 0.10 |
ENSDART00000045684
|
porcn
|
porcupine O-acyltransferase |
| chr15_-_31419805 | 0.10 |
ENSDART00000060111
|
or111-11
|
odorant receptor, family D, subfamily 111, member 11 |
| chr1_+_26667872 | 0.10 |
ENSDART00000152803
ENSDART00000152144 ENSDART00000152785 ENSDART00000152393 |
hemgn
|
hemogen |
| chr14_-_24001825 | 0.08 |
ENSDART00000130704
|
il4
|
interleukin 4 |
| chr6_-_31364475 | 0.07 |
ENSDART00000145715
ENSDART00000134370 |
ak4
|
adenylate kinase 4 |
| chr7_+_22705656 | 0.07 |
ENSDART00000193008
|
si:dkey-165a24.9
|
si:dkey-165a24.9 |
| chr20_+_46620374 | 0.06 |
ENSDART00000005548
|
batf
|
basic leucine zipper transcription factor, ATF-like |
| chr1_-_33353778 | 0.05 |
ENSDART00000041191
|
gyg2
|
glycogenin 2 |
| chr15_-_34878388 | 0.04 |
ENSDART00000154094
|
mgat1a
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase a |
| chr12_+_804314 | 0.03 |
ENSDART00000082169
|
wfikkn2b
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2b |
| chr25_-_19219807 | 0.03 |
ENSDART00000183577
|
acanb
|
aggrecan b |
| chr8_+_21225064 | 0.03 |
ENSDART00000129210
|
cry1ba
|
cryptochrome circadian clock 1ba |
| chr20_+_46202188 | 0.02 |
ENSDART00000100523
|
taar13c
|
trace amine associated receptor 13c |
| chr4_+_7876197 | 0.02 |
ENSDART00000111986
ENSDART00000189601 |
cdc123
|
cell division cycle 123 homolog (S. cerevisiae) |
| chr3_-_26190804 | 0.01 |
ENSDART00000136001
|
ypel3
|
yippee-like 3 |
| chr18_+_6162194 | 0.01 |
ENSDART00000177153
|
otogl
|
otogelin-like |
| chr15_+_17100697 | 0.00 |
ENSDART00000183565
ENSDART00000123197 |
relb
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr1d2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.9 | GO:0034653 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.5 | 4.8 | GO:0046247 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.5 | 2.1 | GO:0006554 | lysine catabolic process(GO:0006554) L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.5 | 7.4 | GO:0090382 | phagosome maturation(GO:0090382) |
| 0.2 | 1.0 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 0.2 | 1.9 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.2 | 1.7 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.2 | 1.3 | GO:0042983 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.2 | 3.0 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.2 | 0.5 | GO:0015882 | L-ascorbic acid transport(GO:0015882) L-ascorbic acid metabolic process(GO:0019852) |
| 0.2 | 0.8 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.1 | 0.4 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 | 1.3 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.1 | 2.1 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 1.3 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.1 | 0.5 | GO:0070376 | ERK5 cascade(GO:0070375) regulation of ERK5 cascade(GO:0070376) positive regulation of ERK5 cascade(GO:0070378) |
| 0.1 | 1.0 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.1 | 0.6 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.3 | GO:0032370 | positive regulation of lipid transport(GO:0032370) |
| 0.1 | 0.4 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 2.8 | GO:0050879 | multicellular organismal movement(GO:0050879) musculoskeletal movement(GO:0050881) |
| 0.1 | 3.4 | GO:0030641 | regulation of cellular pH(GO:0030641) |
| 0.1 | 1.0 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.1 | 3.0 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.1 | 2.1 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.1 | 1.0 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.3 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 1.5 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 0.1 | GO:0061037 | negative regulation of chondrocyte differentiation(GO:0032331) negative regulation of cartilage development(GO:0061037) negative regulation of chondrocyte development(GO:0061182) regulation of bone mineralization involved in bone maturation(GO:1900157) negative regulation of bone mineralization involved in bone maturation(GO:1900158) negative regulation of bone development(GO:1903011) |
| 0.0 | 0.1 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.0 | 0.1 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.0 | 0.4 | GO:0030819 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.0 | 1.1 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.0 | 2.7 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.9 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.1 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 0.0 | 0.7 | GO:0003171 | atrioventricular valve development(GO:0003171) |
| 0.0 | 4.9 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 0.3 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 1.3 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 2.3 | GO:0007249 | I-kappaB kinase/NF-kappaB signaling(GO:0007249) |
| 0.0 | 0.1 | GO:0042832 | response to protozoan(GO:0001562) defense response to protozoan(GO:0042832) |
| 0.0 | 1.1 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 1.6 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 1.5 | GO:0018108 | peptidyl-tyrosine phosphorylation(GO:0018108) |
| 0.0 | 1.0 | GO:0001570 | vasculogenesis(GO:0001570) |
| 0.0 | 0.8 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.7 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.4 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.4 | 5.4 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.2 | 5.7 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 1.7 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 1.6 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 0.4 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 1.0 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 2.0 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 4.0 | GO:0099634 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 2.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.8 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 1.0 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 2.2 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.3 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 5.7 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 4.5 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 2.2 | GO:0045211 | postsynaptic membrane(GO:0045211) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.8 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.5 | 3.0 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.4 | 1.3 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.4 | 2.9 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.3 | 2.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.3 | 1.5 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.3 | 4.9 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 2.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 1.7 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 1.0 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.2 | 0.8 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.1 | 1.0 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 0.4 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 1.0 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.5 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.1 | 1.0 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.1 | 2.7 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 0.3 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 0.3 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.1 | 6.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.8 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 1.0 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.1 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.0 | 3.0 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.1 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.5 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 5.1 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.1 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.4 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.9 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.4 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.9 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 2.2 | GO:0020037 | heme binding(GO:0020037) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.7 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.0 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.2 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.5 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 1.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 1.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 1.0 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.7 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.8 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 2.5 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |