Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
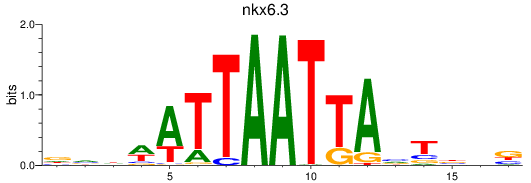
Results for nkx6.3
Z-value: 0.27
Transcription factors associated with nkx6.3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx6.3
|
ENSDARG00000060529 | NK6 homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nkx6.3 | dr11_v1_chr8_+_38415374_38415374 | 0.48 | 3.8e-02 | Click! |
Activity profile of nkx6.3 motif
Sorted Z-values of nkx6.3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_12173554 | 0.53 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr16_+_46111849 | 0.53 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr17_+_15433671 | 0.45 |
ENSDART00000149568
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr17_+_15433518 | 0.45 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr2_+_2223837 | 0.41 |
ENSDART00000101038
ENSDART00000129354 |
tmie
|
transmembrane inner ear |
| chr18_-_17485419 | 0.34 |
ENSDART00000018764
|
foxl1
|
forkhead box L1 |
| chr5_-_30620625 | 0.33 |
ENSDART00000098273
|
tcnl
|
transcobalamin like |
| chr20_+_40457599 | 0.26 |
ENSDART00000017553
|
serinc1
|
serine incorporator 1 |
| chr6_-_37469775 | 0.25 |
ENSDART00000156546
|
plcxd1
|
phosphatidylinositol-specific phospholipase C, X domain containing 1 |
| chr20_-_48485354 | 0.22 |
ENSDART00000124040
ENSDART00000148437 |
insm1a
|
insulinoma-associated 1a |
| chr13_+_31286076 | 0.22 |
ENSDART00000142725
|
gdf10a
|
growth differentiation factor 10a |
| chr9_-_43538328 | 0.21 |
ENSDART00000140526
|
znf385b
|
zinc finger protein 385B |
| chr11_-_2838699 | 0.20 |
ENSDART00000066189
|
lhfpl5a
|
LHFPL tetraspan subfamily member 5a |
| chr8_+_8927870 | 0.20 |
ENSDART00000081985
|
pim2
|
Pim-2 proto-oncogene, serine/threonine kinase |
| chr2_-_7845110 | 0.18 |
ENSDART00000091987
|
si:ch211-38m6.7
|
si:ch211-38m6.7 |
| chr14_+_14662116 | 0.17 |
ENSDART00000161693
|
cetn2
|
centrin, EF-hand protein, 2 |
| chr21_+_34088377 | 0.17 |
ENSDART00000170070
|
mtmr1b
|
myotubularin related protein 1b |
| chr15_+_31344472 | 0.16 |
ENSDART00000146695
ENSDART00000159182 ENSDART00000060125 |
or107-1
|
odorant receptor, family D, subfamily 107, member 1 |
| chr25_-_3058687 | 0.15 |
ENSDART00000149117
ENSDART00000137950 |
si:ch1073-296i8.2
|
si:ch1073-296i8.2 |
| chr20_-_40755614 | 0.15 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr2_+_6243144 | 0.14 |
ENSDART00000058258
|
gng5
|
guanine nucleotide binding protein (G protein), gamma 5 |
| chr11_-_44801968 | 0.13 |
ENSDART00000161846
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr4_-_2637689 | 0.12 |
ENSDART00000192550
ENSDART00000021953 ENSDART00000150344 |
cog5
|
component of oligomeric golgi complex 5 |
| chr20_-_43750771 | 0.12 |
ENSDART00000100605
|
ttc32
|
tetratricopeptide repeat domain 32 |
| chr12_-_10409961 | 0.11 |
ENSDART00000149521
ENSDART00000052001 |
eef2k
|
eukaryotic elongation factor 2 kinase |
| chr11_-_2478374 | 0.11 |
ENSDART00000173205
|
si:ch73-267c23.10
|
si:ch73-267c23.10 |
| chr10_+_2582254 | 0.10 |
ENSDART00000016103
|
nxnl2
|
nucleoredoxin like 2 |
| chr25_-_8660726 | 0.10 |
ENSDART00000168010
ENSDART00000171709 ENSDART00000163426 |
unc45a
|
unc-45 myosin chaperone A |
| chr20_-_45060241 | 0.10 |
ENSDART00000185227
|
klhl29
|
kelch-like family member 29 |
| chr4_+_2637947 | 0.08 |
ENSDART00000130623
|
dus4l
|
dihydrouridine synthase 4-like (S. cerevisiae) |
| chr3_+_18795570 | 0.08 |
ENSDART00000042368
|
fahd1
|
fumarylacetoacetate hydrolase domain containing 1 |
| chr2_-_9818640 | 0.07 |
ENSDART00000139499
ENSDART00000165548 ENSDART00000012442 ENSDART00000046587 |
ap2m1b
|
adaptor-related protein complex 2, mu 1 subunit, b |
| chr11_-_34219211 | 0.06 |
ENSDART00000098472
|
tmem44
|
transmembrane protein 44 |
| chr15_-_43284021 | 0.06 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr16_-_12173399 | 0.06 |
ENSDART00000142574
|
clstn3
|
calsyntenin 3 |
| chr9_-_9415000 | 0.06 |
ENSDART00000146210
|
si:ch211-214p13.9
|
si:ch211-214p13.9 |
| chr21_-_26918901 | 0.05 |
ENSDART00000100685
|
lrfn4a
|
leucine rich repeat and fibronectin type III domain containing 4a |
| chr8_+_50953776 | 0.05 |
ENSDART00000013870
|
zgc:56596
|
zgc:56596 |
| chr3_+_45687266 | 0.05 |
ENSDART00000131652
|
gpr146
|
G protein-coupled receptor 146 |
| chr4_+_4509996 | 0.05 |
ENSDART00000028694
|
gnsa
|
glucosamine (N-acetyl)-6-sulfatase a |
| chr21_+_28445052 | 0.04 |
ENSDART00000077871
|
pygma
|
phosphorylase, glycogen, muscle A |
| chr24_+_16985181 | 0.03 |
ENSDART00000135580
|
eif2s3
|
eukaryotic translation initiation factor 2, subunit 3 gamma |
| chr3_-_26787430 | 0.02 |
ENSDART00000087047
|
rab40c
|
RAB40c, member RAS oncogene family |
| chr23_-_35790235 | 0.01 |
ENSDART00000142369
ENSDART00000141141 ENSDART00000011004 |
mfsd5
|
major facilitator superfamily domain containing 5 |
| chr6_+_54187643 | 0.01 |
ENSDART00000056830
|
pacsin1b
|
protein kinase C and casein kinase substrate in neurons 1b |
| chr3_-_21010979 | 0.01 |
ENSDART00000109578
|
fam117aa
|
family with sequence similarity 117, member Aa |
| chr21_+_43328685 | 0.01 |
ENSDART00000109620
ENSDART00000139668 |
sept8a
|
septin 8a |
| chr23_-_16485190 | 0.00 |
ENSDART00000155038
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx6.3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.2 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.4 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.4 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.2 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 | 0.0 | GO:0052575 | carbohydrate localization(GO:0052575) carbohydrate storage(GO:0052576) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.1 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.2 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |