Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
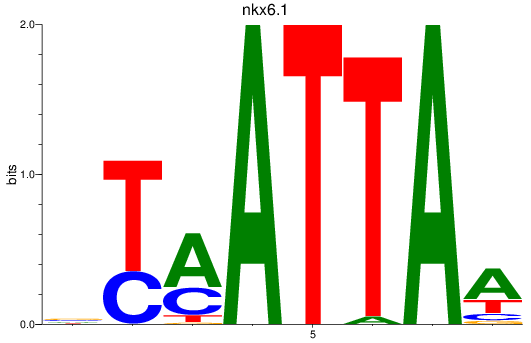
Results for nkx6.1
Z-value: 1.32
Transcription factors associated with nkx6.1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx6.1
|
ENSDARG00000022569 | NK6 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nkx6.1 | dr11_v1_chr21_+_19070921_19070921 | -0.74 | 2.9e-04 | Click! |
Activity profile of nkx6.1 motif
Sorted Z-values of nkx6.1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_30244356 | 10.81 |
ENSDART00000036050
ENSDART00000150080 |
rs1a
|
retinoschisin 1a |
| chr1_-_43905252 | 8.39 |
ENSDART00000135477
ENSDART00000132089 |
si:dkey-22i16.3
|
si:dkey-22i16.3 |
| chr20_-_40755614 | 6.06 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr3_-_50443607 | 5.59 |
ENSDART00000074036
|
rcvrna
|
recoverin a |
| chr25_+_29160102 | 5.56 |
ENSDART00000162854
|
pkmb
|
pyruvate kinase M1/2b |
| chr5_-_30620625 | 5.55 |
ENSDART00000098273
|
tcnl
|
transcobalamin like |
| chr5_-_23362602 | 5.50 |
ENSDART00000137120
|
gria3a
|
glutamate receptor, ionotropic, AMPA 3a |
| chr7_+_14291323 | 4.97 |
ENSDART00000053521
|
rhcga
|
Rh family, C glycoprotein a |
| chr23_+_40460333 | 4.71 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr16_+_46111849 | 4.52 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr18_+_46773198 | 4.24 |
ENSDART00000174647
|
CABZ01041495.1
|
|
| chr15_+_6109861 | 4.20 |
ENSDART00000185154
|
PCP4 (1 of many)
|
Purkinje cell protein 4 |
| chr13_+_29462249 | 3.77 |
ENSDART00000147903
|
lrit1a
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 1a |
| chr17_-_37395460 | 3.76 |
ENSDART00000148160
ENSDART00000075975 |
crip1
|
cysteine-rich protein 1 |
| chr19_+_10339538 | 3.44 |
ENSDART00000151808
ENSDART00000151235 |
rcvrn3
|
recoverin 3 |
| chr5_-_44829719 | 3.44 |
ENSDART00000019104
|
fbp2
|
fructose-1,6-bisphosphatase 2 |
| chr1_-_43915423 | 3.30 |
ENSDART00000181915
ENSDART00000113673 |
scpp5
|
secretory calcium-binding phosphoprotein 5 |
| chr14_+_49135264 | 3.15 |
ENSDART00000084119
|
si:ch1073-44g3.1
|
si:ch1073-44g3.1 |
| chr17_-_16965809 | 3.14 |
ENSDART00000153697
|
nrxn3a
|
neurexin 3a |
| chr9_+_34641237 | 3.08 |
ENSDART00000133996
|
shox
|
short stature homeobox |
| chr10_-_17159761 | 3.00 |
ENSDART00000080449
|
slc5a1
|
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
| chr5_-_38155005 | 2.98 |
ENSDART00000097770
|
gucy2d
|
guanylate cyclase 2D, retinal |
| chr17_-_12389259 | 2.97 |
ENSDART00000185724
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr8_-_25329967 | 2.95 |
ENSDART00000139682
|
eps8l3b
|
EPS8-like 3b |
| chr9_+_33357011 | 2.93 |
ENSDART00000088569
|
nyx
|
nyctalopin |
| chr1_-_22512063 | 2.91 |
ENSDART00000031546
ENSDART00000190987 |
chrna6
|
cholinergic receptor, nicotinic, alpha 6 |
| chr16_-_28658341 | 2.88 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr1_-_23308225 | 2.85 |
ENSDART00000137567
ENSDART00000008201 |
smim14
|
small integral membrane protein 14 |
| chr12_+_24342303 | 2.76 |
ENSDART00000111239
|
nrxn1a
|
neurexin 1a |
| chr22_+_28320792 | 2.75 |
ENSDART00000160562
ENSDART00000164138 |
impg2b
impg2b
|
interphotoreceptor matrix proteoglycan 2b interphotoreceptor matrix proteoglycan 2b |
| chr20_+_18551657 | 2.74 |
ENSDART00000147001
|
si:dkeyp-72h1.1
|
si:dkeyp-72h1.1 |
| chr3_+_1015867 | 2.69 |
ENSDART00000109912
|
si:ch1073-464p5.5
|
si:ch1073-464p5.5 |
| chr10_+_25726694 | 2.68 |
ENSDART00000140308
|
ugt5d1
|
UDP glucuronosyltransferase 5 family, polypeptide D1 |
| chr19_-_6988837 | 2.61 |
ENSDART00000145741
ENSDART00000167640 |
znf384l
|
zinc finger protein 384 like |
| chr16_+_5774977 | 2.61 |
ENSDART00000134202
|
ccka
|
cholecystokinin a |
| chr20_+_4060839 | 2.61 |
ENSDART00000178565
|
TRIM67
|
tripartite motif containing 67 |
| chr21_-_42100471 | 2.60 |
ENSDART00000166148
|
gabra1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr2_-_27329214 | 2.54 |
ENSDART00000145835
|
tmx3a
|
thioredoxin related transmembrane protein 3a |
| chr13_+_23988442 | 2.52 |
ENSDART00000010918
|
agt
|
angiotensinogen |
| chr11_+_13223625 | 2.50 |
ENSDART00000161275
|
abcb11b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11b |
| chr14_-_2933185 | 2.49 |
ENSDART00000161677
ENSDART00000162446 ENSDART00000109378 |
si:dkey-201i24.6
|
si:dkey-201i24.6 |
| chr10_+_29698467 | 2.49 |
ENSDART00000163402
|
dlg2
|
discs, large homolog 2 (Drosophila) |
| chr16_-_45178430 | 2.38 |
ENSDART00000165186
|
si:dkey-33i11.9
|
si:dkey-33i11.9 |
| chr6_-_12588044 | 2.38 |
ENSDART00000047896
|
slc15a1b
|
solute carrier family 15 (oligopeptide transporter), member 1b |
| chr5_-_50992690 | 2.35 |
ENSDART00000149553
ENSDART00000097460 ENSDART00000192021 |
hmgcra
|
3-hydroxy-3-methylglutaryl-CoA reductase a |
| chr10_+_22381802 | 2.35 |
ENSDART00000112484
|
nlgn2b
|
neuroligin 2b |
| chr23_+_20563779 | 2.31 |
ENSDART00000146008
|
camkvl
|
CaM kinase-like vesicle-associated, like |
| chr14_+_50770537 | 2.30 |
ENSDART00000158723
|
sncb
|
synuclein, beta |
| chr5_+_63668735 | 2.28 |
ENSDART00000134261
ENSDART00000097330 |
dnm1b
|
dynamin 1b |
| chr1_-_50859053 | 2.28 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr18_+_9637744 | 2.26 |
ENSDART00000190171
|
pclob
|
piccolo presynaptic cytomatrix protein b |
| chr15_+_36457888 | 2.26 |
ENSDART00000155100
|
si:dkey-262k9.2
|
si:dkey-262k9.2 |
| chr21_+_13366353 | 2.26 |
ENSDART00000151630
|
si:ch73-62l21.1
|
si:ch73-62l21.1 |
| chr22_+_35068046 | 2.25 |
ENSDART00000161660
ENSDART00000169573 |
si:ch73-173h19.3
|
si:ch73-173h19.3 |
| chr18_+_15644559 | 2.24 |
ENSDART00000061794
|
nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr3_-_41791178 | 2.21 |
ENSDART00000049687
|
grifin
|
galectin-related inter-fiber protein |
| chr13_+_38430466 | 2.16 |
ENSDART00000132691
|
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr4_+_7391400 | 2.16 |
ENSDART00000169111
ENSDART00000186395 |
tnni4a
|
troponin I4a |
| chr20_-_40754794 | 2.16 |
ENSDART00000187251
|
cx32.3
|
connexin 32.3 |
| chr17_+_15433518 | 2.14 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr2_+_37227011 | 2.05 |
ENSDART00000126587
ENSDART00000084958 |
samd7
|
sterile alpha motif domain containing 7 |
| chr15_-_16098531 | 2.05 |
ENSDART00000080377
|
aldoca
|
aldolase C, fructose-bisphosphate, a |
| chr7_-_24699985 | 2.04 |
ENSDART00000052802
|
calb2b
|
calbindin 2b |
| chr20_-_29420713 | 2.04 |
ENSDART00000147464
|
ryr3
|
ryanodine receptor 3 |
| chr12_+_31713239 | 2.03 |
ENSDART00000122379
|
habp2
|
hyaluronan binding protein 2 |
| chr14_+_5936996 | 2.02 |
ENSDART00000097144
ENSDART00000126777 |
kctd8
|
potassium channel tetramerization domain containing 8 |
| chr17_+_30448452 | 1.98 |
ENSDART00000153939
|
lpin1
|
lipin 1 |
| chr10_-_24371312 | 1.91 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr6_-_41229787 | 1.90 |
ENSDART00000065013
|
synpr
|
synaptoporin |
| chr17_+_15433671 | 1.87 |
ENSDART00000149568
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr21_+_6780340 | 1.87 |
ENSDART00000139493
ENSDART00000140478 |
olfm1b
|
olfactomedin 1b |
| chr12_-_26153101 | 1.84 |
ENSDART00000076051
|
opn4b
|
opsin 4b |
| chr12_-_35054354 | 1.84 |
ENSDART00000075351
|
zgc:112285
|
zgc:112285 |
| chr3_+_23092762 | 1.84 |
ENSDART00000142884
ENSDART00000024136 |
gngt2a
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2a |
| chr20_+_31076488 | 1.81 |
ENSDART00000136255
ENSDART00000008840 |
otofa
|
otoferlin a |
| chr8_+_24861264 | 1.81 |
ENSDART00000099607
|
slc6a17
|
solute carrier family 6 (neutral amino acid transporter), member 17 |
| chr4_+_12612723 | 1.81 |
ENSDART00000133767
|
lmo3
|
LIM domain only 3 |
| chr16_+_20161805 | 1.81 |
ENSDART00000192146
|
c16h2orf66
|
chromosome 16 C2orf66 homolog |
| chr14_-_4556896 | 1.81 |
ENSDART00000044678
ENSDART00000192863 |
GABRA2
|
gamma-aminobutyric acid type A receptor alpha2 subunit |
| chr21_+_28958471 | 1.80 |
ENSDART00000144331
ENSDART00000005929 |
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr24_+_5208171 | 1.79 |
ENSDART00000155926
ENSDART00000154464 |
si:ch73-206p6.1
|
si:ch73-206p6.1 |
| chr7_+_21841037 | 1.79 |
ENSDART00000077503
|
tm4sf5
|
transmembrane 4 L six family member 5 |
| chr22_+_8753365 | 1.75 |
ENSDART00000106086
|
si:dkey-182g1.2
|
si:dkey-182g1.2 |
| chr8_-_46894362 | 1.74 |
ENSDART00000111124
|
acot7
|
acyl-CoA thioesterase 7 |
| chr12_-_35787801 | 1.74 |
ENSDART00000171682
|
aatkb
|
apoptosis-associated tyrosine kinase b |
| chr5_+_20147830 | 1.73 |
ENSDART00000098727
|
svopa
|
SV2 related protein a |
| chr21_+_8427059 | 1.72 |
ENSDART00000143151
|
dennd1a
|
DENN/MADD domain containing 1A |
| chr2_+_50608099 | 1.71 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr1_+_33969015 | 1.70 |
ENSDART00000042984
ENSDART00000146530 |
epha6
|
eph receptor A6 |
| chr2_+_20332044 | 1.70 |
ENSDART00000112131
|
plppr4a
|
phospholipid phosphatase related 4a |
| chr4_+_7391110 | 1.69 |
ENSDART00000160708
ENSDART00000187823 |
tnni4a
|
troponin I4a |
| chr4_+_21129752 | 1.65 |
ENSDART00000169764
|
syt1a
|
synaptotagmin Ia |
| chr20_-_39271844 | 1.65 |
ENSDART00000192708
|
clu
|
clusterin |
| chr7_+_25059845 | 1.63 |
ENSDART00000077215
|
ppp2r5b
|
protein phosphatase 2, regulatory subunit B', beta |
| chr2_-_37465517 | 1.63 |
ENSDART00000139983
|
si:dkey-57k2.6
|
si:dkey-57k2.6 |
| chr16_+_13818500 | 1.62 |
ENSDART00000135245
|
flcn
|
folliculin |
| chr16_-_52540056 | 1.61 |
ENSDART00000188304
|
CR293507.1
|
|
| chr15_+_21276735 | 1.61 |
ENSDART00000111213
|
ubash3bb
|
ubiquitin associated and SH3 domain containing Bb |
| chr9_+_54417141 | 1.61 |
ENSDART00000056810
|
drd1b
|
dopamine receptor D1b |
| chr16_+_34531486 | 1.59 |
ENSDART00000043291
|
paqr7b
|
progestin and adipoQ receptor family member VII, b |
| chr3_-_13146631 | 1.59 |
ENSDART00000172460
|
prkar1b
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr5_+_36611128 | 1.59 |
ENSDART00000097684
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr16_+_2820340 | 1.57 |
ENSDART00000092299
ENSDART00000192931 ENSDART00000148512 |
si:dkey-288i20.2
|
si:dkey-288i20.2 |
| chr5_-_43071058 | 1.56 |
ENSDART00000165546
|
si:dkey-245n4.2
|
si:dkey-245n4.2 |
| chr11_-_6188413 | 1.56 |
ENSDART00000109972
|
ccl44
|
chemokine (C-C motif) ligand 44 |
| chr7_-_28147838 | 1.55 |
ENSDART00000158921
|
lmo1
|
LIM domain only 1 |
| chr7_-_28148310 | 1.55 |
ENSDART00000044208
|
lmo1
|
LIM domain only 1 |
| chr9_-_43538328 | 1.55 |
ENSDART00000140526
|
znf385b
|
zinc finger protein 385B |
| chr13_+_38521152 | 1.54 |
ENSDART00000145292
|
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr4_-_8903240 | 1.54 |
ENSDART00000129983
|
mpped1
|
metallophosphoesterase domain containing 1 |
| chr7_+_38510197 | 1.53 |
ENSDART00000173468
ENSDART00000100479 |
slc7a9
|
solute carrier family 7 (amino acid transporter light chain, bo,+ system), member 9 |
| chr19_-_205104 | 1.53 |
ENSDART00000011890
|
zbtb22a
|
zinc finger and BTB domain containing 22a |
| chr24_-_38110779 | 1.52 |
ENSDART00000147783
|
crp
|
c-reactive protein, pentraxin-related |
| chr10_+_32683089 | 1.52 |
ENSDART00000063551
|
ppm1e
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E |
| chr22_-_13851297 | 1.51 |
ENSDART00000080306
|
s100b
|
S100 calcium binding protein, beta (neural) |
| chr6_-_38419318 | 1.47 |
ENSDART00000138026
|
gabra5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| chr22_+_8753092 | 1.46 |
ENSDART00000140720
|
si:dkey-182g1.2
|
si:dkey-182g1.2 |
| chr13_-_45475289 | 1.46 |
ENSDART00000043345
|
rsrp1
|
arginine/serine-rich protein 1 |
| chr7_+_25033924 | 1.45 |
ENSDART00000170873
|
sb:cb1058
|
sb:cb1058 |
| chr19_-_5103313 | 1.45 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr21_-_40174647 | 1.45 |
ENSDART00000183738
ENSDART00000076840 ENSDART00000145109 |
slco2b1
|
solute carrier organic anion transporter family, member 2B1 |
| chr23_-_306796 | 1.45 |
ENSDART00000143125
|
anks1aa
|
ankyrin repeat and sterile alpha motif domain containing 1Aa |
| chr3_-_28258462 | 1.43 |
ENSDART00000191573
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr18_+_10884996 | 1.42 |
ENSDART00000147613
|
mical3a
|
microtubule associated monooxygenase, calponin and LIM domain containing 3a |
| chr3_+_29714775 | 1.42 |
ENSDART00000041388
|
cacng2a
|
calcium channel, voltage-dependent, gamma subunit 2a |
| chr1_+_8601935 | 1.42 |
ENSDART00000152367
|
si:ch211-160d14.6
|
si:ch211-160d14.6 |
| chr20_+_25879826 | 1.41 |
ENSDART00000018519
|
zgc:153896
|
zgc:153896 |
| chr12_+_18681477 | 1.40 |
ENSDART00000127981
ENSDART00000143979 |
rgs9b
|
regulator of G protein signaling 9b |
| chr5_+_51443009 | 1.39 |
ENSDART00000083350
|
rasgrf2b
|
Ras protein-specific guanine nucleotide-releasing factor 2b |
| chr19_-_6631900 | 1.39 |
ENSDART00000144571
|
pvrl2l
|
poliovirus receptor-related 2 like |
| chr8_-_13184989 | 1.38 |
ENSDART00000135738
|
zgc:194990
|
zgc:194990 |
| chr11_-_37509001 | 1.38 |
ENSDART00000109753
|
bsnb
|
bassoon (presynaptic cytomatrix protein) b |
| chr22_-_15010688 | 1.37 |
ENSDART00000139892
|
elfn2a
|
extracellular leucine-rich repeat and fibronectin type III domain containing 2a |
| chr2_+_10127762 | 1.36 |
ENSDART00000100726
|
insl5b
|
insulin-like 5b |
| chr25_-_4313699 | 1.36 |
ENSDART00000154038
|
syt7a
|
synaptotagmin VIIa |
| chr8_-_14126646 | 1.35 |
ENSDART00000027225
|
bgna
|
biglycan a |
| chr3_+_33341640 | 1.34 |
ENSDART00000186352
|
pyya
|
peptide YYa |
| chr16_-_38118003 | 1.34 |
ENSDART00000058667
|
si:dkey-23o4.6
|
si:dkey-23o4.6 |
| chr6_-_10320676 | 1.34 |
ENSDART00000151247
|
scn1lab
|
sodium channel, voltage-gated, type I like, alpha b |
| chr3_+_17537352 | 1.33 |
ENSDART00000104549
|
hcrt
|
hypocretin (orexin) neuropeptide precursor |
| chr23_-_38497705 | 1.33 |
ENSDART00000109493
|
tshz2
|
teashirt zinc finger homeobox 2 |
| chr15_-_6247775 | 1.32 |
ENSDART00000148350
|
dscamb
|
Down syndrome cell adhesion molecule b |
| chr1_+_11977426 | 1.31 |
ENSDART00000103399
|
tspan5b
|
tetraspanin 5b |
| chr17_-_19019635 | 1.30 |
ENSDART00000126666
|
flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr5_-_6567464 | 1.30 |
ENSDART00000184985
|
tnks1bp1
|
tankyrase 1 binding protein 1 |
| chr23_-_1348933 | 1.28 |
ENSDART00000168981
|
CABZ01078120.1
|
|
| chr3_-_33395233 | 1.28 |
ENSDART00000167349
|
si:dkey-283b1.6
|
si:dkey-283b1.6 |
| chr1_-_43899422 | 1.28 |
ENSDART00000134649
|
si:dkey-22i16.2
|
si:dkey-22i16.2 |
| chr23_-_32162810 | 1.26 |
ENSDART00000155905
|
nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr15_+_22311803 | 1.26 |
ENSDART00000150182
|
hepacama
|
hepatic and glial cell adhesion molecule a |
| chr12_-_19007834 | 1.26 |
ENSDART00000153248
|
chadlb
|
chondroadherin-like b |
| chr3_+_28953274 | 1.26 |
ENSDART00000133528
ENSDART00000103602 |
lgals2a
|
lectin, galactoside-binding, soluble, 2a |
| chr1_+_52929185 | 1.26 |
ENSDART00000147683
|
inpp4b
|
inositol polyphosphate-4-phosphatase type II B |
| chr6_+_23887314 | 1.26 |
ENSDART00000163188
|
znf648
|
zinc finger protein 648 |
| chr5_-_25174420 | 1.25 |
ENSDART00000141554
|
abca2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr3_+_34919810 | 1.25 |
ENSDART00000055264
|
ca10b
|
carbonic anhydrase Xb |
| chr10_+_41765616 | 1.25 |
ENSDART00000170682
|
rnf34b
|
ring finger protein 34b |
| chr16_+_17389116 | 1.25 |
ENSDART00000103750
ENSDART00000173448 |
fam131bb
|
family with sequence similarity 131, member Bb |
| chr1_+_52392511 | 1.24 |
ENSDART00000144025
|
si:ch211-217k17.8
|
si:ch211-217k17.8 |
| chr4_+_11384891 | 1.24 |
ENSDART00000092381
ENSDART00000186577 ENSDART00000191054 ENSDART00000191584 |
pcloa
|
piccolo presynaptic cytomatrix protein a |
| chr1_-_5455498 | 1.23 |
ENSDART00000040368
ENSDART00000114035 |
mnx2b
|
motor neuron and pancreas homeobox 2b |
| chr13_-_33170733 | 1.22 |
ENSDART00000057382
|
fbln5
|
fibulin 5 |
| chr24_-_1985007 | 1.21 |
ENSDART00000189870
|
PARD3 (1 of many)
|
par-3 family cell polarity regulator |
| chr1_-_19215336 | 1.19 |
ENSDART00000162949
ENSDART00000170680 |
ptprdb
|
protein tyrosine phosphatase, receptor type, D, b |
| chr23_+_21638258 | 1.19 |
ENSDART00000104188
|
igsf21b
|
immunoglobin superfamily, member 21b |
| chr19_-_5103141 | 1.19 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr9_+_52398531 | 1.19 |
ENSDART00000126215
|
dap1b
|
death associated protein 1b |
| chr10_-_13343831 | 1.19 |
ENSDART00000135941
|
il11ra
|
interleukin 11 receptor, alpha |
| chr10_-_8060573 | 1.18 |
ENSDART00000147104
ENSDART00000099030 |
si:ch211-251f6.6
|
si:ch211-251f6.6 |
| chr1_-_12160981 | 1.18 |
ENSDART00000162146
|
b4galt1
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 |
| chr19_-_31522625 | 1.17 |
ENSDART00000158438
ENSDART00000035049 |
necab1
|
N-terminal EF-hand calcium binding protein 1 |
| chr8_-_20230559 | 1.16 |
ENSDART00000193677
|
mllt1a
|
MLLT1, super elongation complex subunit a |
| chr16_-_17162843 | 1.16 |
ENSDART00000089386
|
iffo1b
|
intermediate filament family orphan 1b |
| chr20_-_2949028 | 1.16 |
ENSDART00000104667
ENSDART00000193151 ENSDART00000131946 |
cdk19
|
cyclin-dependent kinase 19 |
| chr21_+_17768174 | 1.15 |
ENSDART00000141380
|
rxraa
|
retinoid X receptor, alpha a |
| chr21_-_22827548 | 1.15 |
ENSDART00000079161
|
angptl5
|
angiopoietin-like 5 |
| chr23_+_28731379 | 1.14 |
ENSDART00000047378
|
cort
|
cortistatin |
| chr24_+_13316737 | 1.14 |
ENSDART00000191658
|
SBSPON
|
somatomedin B and thrombospondin type 1 domain containing |
| chr2_+_6963296 | 1.13 |
ENSDART00000147146
|
ddr2b
|
discoidin domain receptor tyrosine kinase 2b |
| chr13_+_25449681 | 1.12 |
ENSDART00000101328
|
atoh7
|
atonal bHLH transcription factor 7 |
| chr3_-_35800221 | 1.11 |
ENSDART00000031390
|
caskin1
|
CASK interacting protein 1 |
| chr7_-_69521481 | 1.11 |
ENSDART00000148465
|
slc1a1
|
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 |
| chr23_-_20051369 | 1.10 |
ENSDART00000049836
|
bgnb
|
biglycan b |
| chr6_-_35779348 | 1.10 |
ENSDART00000191159
|
brinp3a.1
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3a, tandem duplicate 1 |
| chr10_+_37137464 | 1.09 |
ENSDART00000114909
|
cuedc1a
|
CUE domain containing 1a |
| chr9_+_31795343 | 1.09 |
ENSDART00000139584
|
itgbl1
|
integrin, beta-like 1 |
| chr23_-_35790235 | 1.09 |
ENSDART00000142369
ENSDART00000141141 ENSDART00000011004 |
mfsd5
|
major facilitator superfamily domain containing 5 |
| chr1_-_45042210 | 1.08 |
ENSDART00000073694
|
smu1b
|
SMU1, DNA replication regulator and spliceosomal factor b |
| chr16_+_23431189 | 1.08 |
ENSDART00000004679
|
icn
|
ictacalcin |
| chr25_+_24291156 | 1.07 |
ENSDART00000083407
|
b4galnt4a
|
beta-1,4-N-acetyl-galactosaminyl transferase 4a |
| chr19_-_7358184 | 1.07 |
ENSDART00000092379
|
oxr1b
|
oxidation resistance 1b |
| chr7_+_71547747 | 1.07 |
ENSDART00000180869
|
adcyap1a
|
adenylate cyclase activating polypeptide 1a |
| chr13_+_4225173 | 1.07 |
ENSDART00000058242
ENSDART00000143456 |
mea1
|
male-enhanced antigen 1 |
| chr23_+_20110086 | 1.06 |
ENSDART00000054664
|
tnnc1b
|
troponin C type 1b (slow) |
| chr19_+_42470396 | 1.06 |
ENSDART00000191679
|
si:dkey-166k12.1
|
si:dkey-166k12.1 |
| chr8_-_34052019 | 1.06 |
ENSDART00000040126
ENSDART00000159208 ENSDART00000048994 ENSDART00000098822 |
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr11_+_28476298 | 1.06 |
ENSDART00000122319
|
lrrc38b
|
leucine rich repeat containing 38b |
| chr14_-_1454045 | 1.05 |
ENSDART00000161460
|
pmt
|
phosphoethanolamine methyltransferase |
| chr7_+_17229980 | 1.05 |
ENSDART00000184910
|
slc6a5
|
solute carrier family 6 (neurotransmitter transporter), member 5 |
| chr19_+_5480327 | 1.04 |
ENSDART00000148794
|
jupb
|
junction plakoglobin b |
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx6.1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:0071236 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 1.1 | 3.4 | GO:0005985 | sucrose metabolic process(GO:0005985) sucrose biosynthetic process(GO:0005986) disaccharide biosynthetic process(GO:0046351) |
| 0.8 | 9.0 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.7 | 3.7 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.7 | 2.6 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.6 | 2.5 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.6 | 2.5 | GO:0015722 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.6 | 5.0 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.6 | 1.8 | GO:0015824 | proline transport(GO:0015824) |
| 0.5 | 2.6 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.5 | 2.0 | GO:1900271 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.5 | 2.9 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.5 | 4.9 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.5 | 1.5 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.4 | 1.3 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.3 | 1.3 | GO:0042755 | eating behavior(GO:0042755) |
| 0.3 | 0.9 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.3 | 2.2 | GO:2001271 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.3 | 0.9 | GO:0021611 | facial nerve formation(GO:0021611) |
| 0.3 | 1.7 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.3 | 2.8 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.3 | 2.4 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.2 | 0.7 | GO:0015942 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.2 | 0.7 | GO:0052575 | carbohydrate localization(GO:0052575) carbohydrate storage(GO:0052576) |
| 0.2 | 1.0 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.2 | 0.9 | GO:0099543 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.2 | 1.6 | GO:0042311 | vasodilation(GO:0042311) |
| 0.2 | 2.3 | GO:0097105 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.2 | 3.0 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.2 | 1.0 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.2 | 0.6 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.2 | 1.4 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.2 | 1.0 | GO:0039528 | cytoplasmic pattern recognition receptor signaling pathway(GO:0002753) cytoplasmic pattern recognition receptor signaling pathway in response to virus(GO:0039528) cellular response to virus(GO:0098586) |
| 0.2 | 3.7 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.2 | 0.8 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.2 | 1.2 | GO:0042983 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.2 | 1.3 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.2 | 0.8 | GO:1902514 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.1 | 1.0 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 2.7 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.1 | 1.3 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 2.2 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.1 | 1.8 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.1 | 1.1 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 0.5 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.1 | 1.0 | GO:0043092 | amino acid import(GO:0043090) L-amino acid import(GO:0043092) |
| 0.1 | 1.0 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.1 | 0.4 | GO:1901890 | positive regulation of cell junction assembly(GO:1901890) |
| 0.1 | 1.8 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 2.2 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 3.0 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 0.3 | GO:2000639 | response to sterol(GO:0036314) cellular response to sterol(GO:0036315) SREBP-SCAP complex retention in endoplasmic reticulum(GO:0036316) regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 | 5.9 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 2.5 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 2.9 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.1 | 0.3 | GO:0019878 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.1 | 4.0 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.1 | 0.8 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 1.0 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) |
| 0.1 | 0.7 | GO:0070285 | pigment cell development(GO:0070285) |
| 0.1 | 0.8 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 1.6 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 7.3 | GO:0032869 | cellular response to insulin stimulus(GO:0032869) |
| 0.1 | 2.0 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 1.0 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 0.7 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.1 | 2.9 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.1 | 1.4 | GO:0015858 | nucleoside transport(GO:0015858) nucleoside transmembrane transport(GO:1901642) |
| 0.1 | 0.7 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.1 | 0.4 | GO:0016038 | absorption of visible light(GO:0016038) |
| 0.1 | 1.6 | GO:0001990 | regulation of systemic arterial blood pressure by hormone(GO:0001990) |
| 0.1 | 1.3 | GO:0090303 | positive regulation of wound healing(GO:0090303) |
| 0.1 | 0.9 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.1 | 0.4 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.7 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 1.7 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 | 0.8 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 | 1.5 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.1 | 2.3 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.1 | 1.0 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.1 | 0.2 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.1 | 1.0 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 2.6 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 0.6 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.1 | 1.6 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.1 | 0.2 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.1 | 2.6 | GO:0007586 | digestion(GO:0007586) |
| 0.1 | 1.7 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.1 | 1.3 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.1 | 2.3 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 2.4 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 1.0 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 0.5 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 0.2 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.1 | 1.2 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.1 | 2.0 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.1 | 1.1 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.1 | 2.3 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.1 | 1.3 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 1.0 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 2.2 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 0.4 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 0.3 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.0 | 0.1 | GO:0070589 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.0 | 0.8 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 3.2 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 0.5 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.8 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 1.5 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.4 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.7 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 1.4 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 0.2 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.0 | 0.4 | GO:0046247 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.0 | 0.8 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.6 | GO:0048512 | rhythmic behavior(GO:0007622) circadian behavior(GO:0048512) |
| 0.0 | 5.5 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 0.8 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.7 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.5 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 1.1 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.7 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 1.6 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 1.9 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.4 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 3.0 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.5 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 0.7 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.0 | 1.5 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 0.4 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 0.2 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.8 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 1.5 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.7 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.1 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.0 | 0.3 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 1.3 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.7 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 3.7 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 0.2 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 2.4 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 0.3 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 1.1 | GO:0070374 | positive regulation of ERK1 and ERK2 cascade(GO:0070374) |
| 0.0 | 0.3 | GO:0098969 | neurotransmitter receptor transport to plasma membrane(GO:0098877) neurotransmitter receptor transport to postsynaptic membrane(GO:0098969) establishment of protein localization to postsynaptic membrane(GO:1903540) |
| 0.0 | 4.0 | GO:0042391 | regulation of membrane potential(GO:0042391) |
| 0.0 | 0.7 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 1.5 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.0 | 0.8 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
| 0.0 | 0.6 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.1 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.0 | 0.6 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.6 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 1.0 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.7 | GO:1903844 | regulation of transforming growth factor beta receptor signaling pathway(GO:0017015) regulation of cellular response to transforming growth factor beta stimulus(GO:1903844) |
| 0.0 | 0.9 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 1.3 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.7 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.3 | GO:0071875 | adrenergic receptor signaling pathway(GO:0071875) |
| 0.0 | 0.1 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 1.7 | GO:0015698 | inorganic anion transport(GO:0015698) |
| 0.0 | 0.5 | GO:1990573 | potassium ion import(GO:0010107) potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.5 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0070255 | regulation of mucus secretion(GO:0070255) positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.2 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.5 | GO:0043648 | dicarboxylic acid metabolic process(GO:0043648) |
| 0.0 | 0.9 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.0 | 2.0 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.9 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.0 | 1.4 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.6 | 3.7 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.5 | 3.0 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.5 | 4.9 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.3 | 2.8 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.3 | 1.7 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.3 | 1.7 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.3 | 1.8 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 3.7 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.2 | 1.3 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.2 | 1.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.2 | 0.5 | GO:0030062 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.2 | 0.6 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.2 | 6.9 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 5.0 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 4.3 | GO:0043679 | axon terminus(GO:0043679) neuron projection terminus(GO:0044306) |
| 0.1 | 2.4 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 5.9 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 2.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 1.2 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 0.5 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 9.0 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 0.3 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.8 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.7 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 0.4 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 3.0 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 1.7 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.7 | GO:0038039 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.1 | 0.3 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 1.6 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 1.3 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.1 | 0.7 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 2.8 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 0.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 1.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.1 | 1.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.4 | GO:0071914 | prominosome(GO:0071914) |
| 0.0 | 0.7 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.9 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 2.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.1 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 2.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 3.2 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 1.3 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.6 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 1.4 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 5.8 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 3.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.8 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 1.1 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 1.0 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.4 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.9 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.6 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.9 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 3.1 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.8 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 2.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.8 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 0.1 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 20.2 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.3 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 1.6 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.2 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 3.3 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 15.8 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.4 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 1.2 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.1 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.4 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 1.0 | 2.9 | GO:0015462 | protein-transmembrane transporting ATPase activity(GO:0015462) |
| 0.9 | 5.6 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.8 | 2.3 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.7 | 2.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.7 | 2.2 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.7 | 1.4 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.7 | 2.6 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.7 | 2.0 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.6 | 2.5 | GO:0015126 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.6 | 3.0 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.5 | 1.6 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.4 | 1.5 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.4 | 2.2 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.4 | 5.5 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.4 | 2.5 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.4 | 2.8 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.3 | 2.4 | GO:0042936 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.3 | 1.8 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.3 | 1.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.3 | 2.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.3 | 4.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.3 | 1.9 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.3 | 4.9 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.3 | 1.3 | GO:0031841 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.3 | 1.3 | GO:0048763 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.3 | 4.0 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.3 | 1.1 | GO:0016521 | pituitary adenylate cyclase activating polypeptide activity(GO:0016521) |
| 0.3 | 2.0 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.2 | 1.7 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.2 | 1.6 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.2 | 3.9 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.2 | 1.1 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.2 | 1.0 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.2 | 5.0 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.2 | 0.6 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.2 | 0.8 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.2 | 1.0 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.2 | 1.8 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 1.8 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.7 | GO:0005542 | folic acid binding(GO:0005542) ammonia-lyase activity(GO:0016841) |
| 0.1 | 0.7 | GO:0004645 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 0.7 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.8 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 3.1 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 4.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 1.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 1.3 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 1.1 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.1 | 0.4 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.1 | 3.9 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 4.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 3.8 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 1.6 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 0.4 | GO:0047777 | (3S)-citramalyl-CoA lyase activity(GO:0047777) |
| 0.1 | 1.0 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 1.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 2.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.4 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 0.5 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.1 | 0.4 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 0.8 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 2.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.7 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 1.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 0.6 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.1 | 1.9 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 3.2 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 1.0 | GO:0005222 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 1.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 1.0 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 2.1 | GO:0005343 | organic acid:sodium symporter activity(GO:0005343) |
| 0.1 | 1.3 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 2.3 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 1.0 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.1 | 1.7 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 1.0 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.8 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 0.3 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 3.3 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.8 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 0.3 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 1.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.7 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.7 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 1.0 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 2.1 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.9 | GO:0016917 | GABA receptor activity(GO:0016917) |
| 0.0 | 0.7 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 1.6 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.4 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 0.4 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 2.4 | GO:0050661 | NADP binding(GO:0050661) |
| 0.0 | 0.6 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 1.9 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 1.0 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.7 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.5 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.9 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.5 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 1.6 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 1.4 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.2 | GO:0004997 | thyrotropin-releasing hormone receptor activity(GO:0004997) |
| 0.0 | 0.1 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.0 | 0.6 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.8 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 3.6 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.6 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 1.0 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.1 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.0 | 1.9 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 5.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 2.0 | GO:1901981 | phosphatidylinositol phosphate binding(GO:1901981) |
| 0.0 | 1.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.9 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.0 | 0.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 1.5 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.0 | 0.1 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 2.2 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 2.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 1.0 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.3 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.5 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.6 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 2.5 | GO:0015399 | primary active transmembrane transporter activity(GO:0015399) |
| 0.0 | 0.5 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 2.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.5 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.3 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 0.3 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.7 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.2 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 10.3 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.2 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.1 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.6 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.1 | GO:0008199 | ferric iron binding(GO:0008199) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.0 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.2 | 3.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 1.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 1.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 2.4 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 2.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.0 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.8 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.1 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.0 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 2.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.5 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.2 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 2.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 6.8 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.3 | 2.9 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.2 | 1.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 1.6 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.2 | 2.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.2 | 0.9 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.2 | 1.5 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.2 | 1.5 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.2 | 1.1 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.2 | 2.0 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.2 | 2.1 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.1 | 1.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 3.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 0.8 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 3.0 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 0.2 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 1.5 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 0.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.8 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 4.0 | REACTOME TRANSPORT OF GLUCOSE AND OTHER SUGARS BILE SALTS AND ORGANIC ACIDS METAL IONS AND AMINE COMPOUNDS | Genes involved in Transport of glucose and other sugars, bile salts and organic acids, metal ions and amine compounds |
| 0.1 | 1.0 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.1 | 0.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 5.8 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 0.4 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.8 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.6 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 1.3 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 1.0 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 1.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.4 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.3 | REACTOME PI3K EVENTS IN ERBB4 SIGNALING | Genes involved in PI3K events in ERBB4 signaling |
| 0.0 | 0.4 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.3 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |