Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
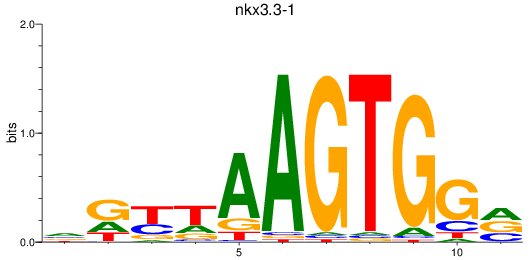
Results for nkx3.3-1
Z-value: 1.10
Transcription factors associated with nkx3.3-1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx3.3-1
|
ENSDARG00000110589 | NK3 homeobox 3 |
Activity profile of nkx3.3-1 motif
Sorted Z-values of nkx3.3-1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_724639 | 2.92 |
ENSDART00000105323
|
zgc:162255
|
zgc:162255 |
| chr19_-_35155722 | 2.10 |
ENSDART00000151924
|
macf1a
|
microtubule-actin crosslinking factor 1a |
| chr2_+_55916911 | 1.62 |
ENSDART00000189483
ENSDART00000183647 ENSDART00000083470 |
atcayb
|
ataxia, cerebellar, Cayman type b |
| chr24_-_33308045 | 1.57 |
ENSDART00000149711
|
slc4a2b
|
solute carrier family 4 (anion exchanger), member 2b |
| chr16_-_13612650 | 1.55 |
ENSDART00000080372
|
dbpb
|
D site albumin promoter binding protein b |
| chr23_+_36601984 | 1.50 |
ENSDART00000128598
|
igfbp6b
|
insulin-like growth factor binding protein 6b |
| chr25_+_28282274 | 1.49 |
ENSDART00000164502
|
aass
|
aminoadipate-semialdehyde synthase |
| chr2_+_39618951 | 1.48 |
ENSDART00000077108
|
zgc:136870
|
zgc:136870 |
| chr7_-_23777445 | 1.48 |
ENSDART00000173527
|
si:ch211-200p22.4
|
si:ch211-200p22.4 |
| chr22_+_7480465 | 1.45 |
ENSDART00000034545
|
CELA1 (1 of many)
|
zgc:92745 |
| chr21_+_5589923 | 1.41 |
ENSDART00000160885
|
stbd1
|
starch binding domain 1 |
| chr9_-_43073960 | 1.38 |
ENSDART00000059460
|
ttn.2
|
titin, tandem duplicate 2 |
| chr20_-_53078607 | 1.33 |
ENSDART00000163494
ENSDART00000191730 |
CABZ01066813.1
|
|
| chr5_+_2815021 | 1.31 |
ENSDART00000020472
|
hpda
|
4-hydroxyphenylpyruvate dioxygenase a |
| chr22_+_19188809 | 1.20 |
ENSDART00000134791
ENSDART00000133682 |
si:dkey-21e2.8
|
si:dkey-21e2.8 |
| chr10_-_33343244 | 1.16 |
ENSDART00000164191
|
c2cd2
|
C2 calcium-dependent domain containing 2 |
| chr1_-_54706039 | 1.15 |
ENSDART00000083633
|
exosc1
|
exosome component 1 |
| chr19_+_43256986 | 1.12 |
ENSDART00000182336
|
DGKD
|
diacylglycerol kinase delta |
| chr19_+_8612839 | 1.11 |
ENSDART00000144925
|
snx27a
|
sorting nexin family member 27a |
| chr20_-_3911546 | 1.05 |
ENSDART00000169787
|
cnksr3
|
cnksr family member 3 |
| chr10_+_25726694 | 1.02 |
ENSDART00000140308
|
ugt5d1
|
UDP glucuronosyltransferase 5 family, polypeptide D1 |
| chr11_-_7050825 | 1.01 |
ENSDART00000179749
|
si:ch211-253b8.5
|
si:ch211-253b8.5 |
| chr9_-_49531762 | 1.00 |
ENSDART00000121875
|
xirp2b
|
xin actin binding repeat containing 2b |
| chr7_-_5487593 | 0.99 |
ENSDART00000136594
|
arhgef11
|
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr16_-_22585289 | 0.99 |
ENSDART00000134239
ENSDART00000193959 ENSDART00000077998 |
si:dkey-238m4.3
cgna
|
si:dkey-238m4.3 cingulin a |
| chr9_-_7390388 | 0.99 |
ENSDART00000132392
|
slc23a3
|
solute carrier family 23, member 3 |
| chr5_-_5243079 | 0.99 |
ENSDART00000130576
ENSDART00000164377 |
mvb12ba
|
multivesicular body subunit 12Ba |
| chr6_+_3280939 | 0.98 |
ENSDART00000151359
|
kdm4aa
|
lysine (K)-specific demethylase 4A, genome duplicate a |
| chr10_+_37268854 | 0.97 |
ENSDART00000131897
|
nf1b
|
neurofibromin 1b |
| chr7_-_32833153 | 0.96 |
ENSDART00000099871
ENSDART00000099872 |
slc17a6b
|
solute carrier family 17 (vesicular glutamate transporter), member 6b |
| chr16_+_38240027 | 0.95 |
ENSDART00000111081
|
prune
|
prune exopolyphosphatase |
| chr20_+_46741074 | 0.95 |
ENSDART00000145294
|
si:ch211-57i17.1
|
si:ch211-57i17.1 |
| chr1_-_11075403 | 0.93 |
ENSDART00000102903
ENSDART00000170290 |
dmd
|
dystrophin |
| chr24_+_15670013 | 0.92 |
ENSDART00000185826
|
CU929414.1
|
|
| chr8_-_51507144 | 0.92 |
ENSDART00000024882
ENSDART00000135166 |
fgfr1a
|
fibroblast growth factor receptor 1a |
| chr16_+_4658250 | 0.92 |
ENSDART00000006212
|
LIN28A
|
si:ch1073-284b18.2 |
| chr21_-_28523548 | 0.92 |
ENSDART00000077910
|
epdl2
|
ependymin-like 2 |
| chr8_+_8712446 | 0.91 |
ENSDART00000158674
|
elk1
|
ELK1, member of ETS oncogene family |
| chr19_-_6840506 | 0.89 |
ENSDART00000081568
|
tcf19l
|
transcription factor 19 (SC1), like |
| chr16_+_3982590 | 0.87 |
ENSDART00000149295
|
zc3h12a
|
zinc finger CCCH-type containing 12A |
| chr2_+_45593783 | 0.85 |
ENSDART00000148037
|
fndc7rs2
|
fibronectin type III domain containing 7, related sequence 2 |
| chr12_-_9617770 | 0.84 |
ENSDART00000106267
|
arg1
|
arginase 1 |
| chr5_-_57820873 | 0.83 |
ENSDART00000089961
|
sik2a
|
salt-inducible kinase 2a |
| chr5_-_57204352 | 0.82 |
ENSDART00000171252
ENSDART00000180727 |
man2a1
|
mannosidase, alpha, class 2A, member 1 |
| chr18_+_26829362 | 0.82 |
ENSDART00000132728
|
slc28a1
|
solute carrier family 28 (concentrative nucleoside transporter), member 1 |
| chr17_-_200316 | 0.81 |
ENSDART00000190561
|
CABZ01083778.1
|
|
| chr4_-_12862087 | 0.80 |
ENSDART00000080536
|
hmga2
|
high mobility group AT-hook 2 |
| chr17_-_16069905 | 0.80 |
ENSDART00000110383
|
map7a
|
microtubule-associated protein 7a |
| chr4_+_61995745 | 0.79 |
ENSDART00000171539
|
CT990567.1
|
|
| chr22_-_20376488 | 0.79 |
ENSDART00000140187
|
zbtb7a
|
zinc finger and BTB domain containing 7a |
| chr1_-_50791280 | 0.78 |
ENSDART00000181224
|
CABZ01031870.1
|
|
| chr21_+_26612777 | 0.78 |
ENSDART00000142667
|
esrra
|
estrogen-related receptor alpha |
| chr15_+_45595385 | 0.78 |
ENSDART00000161937
ENSDART00000170214 ENSDART00000157450 |
atg16l1
|
ATG16 autophagy related 16-like 1 (S. cerevisiae) |
| chr13_-_37109987 | 0.77 |
ENSDART00000136750
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr1_+_7988052 | 0.77 |
ENSDART00000167552
|
CR855320.2
|
|
| chr8_+_25254435 | 0.76 |
ENSDART00000143554
|
ampd2b
|
adenosine monophosphate deaminase 2b |
| chr6_-_1820606 | 0.75 |
ENSDART00000183228
|
FO834857.1
|
|
| chr19_+_7810028 | 0.73 |
ENSDART00000081592
ENSDART00000140719 |
aqp10b
|
aquaporin 10b |
| chr21_+_10021823 | 0.73 |
ENSDART00000163995
|
herc7
|
hect domain and RLD 7 |
| chr10_+_5159475 | 0.71 |
ENSDART00000142507
|
cdc42se2
|
CDC42 small effector 2 |
| chr4_-_8030583 | 0.70 |
ENSDART00000113628
|
si:ch211-240l19.8
|
si:ch211-240l19.8 |
| chr21_-_2162850 | 0.69 |
ENSDART00000159731
|
gb:ai877918
|
expressed sequence AI877918 |
| chr22_-_26236188 | 0.69 |
ENSDART00000162640
ENSDART00000167169 ENSDART00000138595 |
c3b.1
|
complement component c3b, tandem duplicate 1 |
| chr4_-_71708567 | 0.68 |
ENSDART00000182645
|
si:dkeyp-4f2.1
|
si:dkeyp-4f2.1 |
| chr2_+_42177113 | 0.68 |
ENSDART00000056441
|
tmeff1a
|
transmembrane protein with EGF-like and two follistatin-like domains 1a |
| chr9_-_27442339 | 0.67 |
ENSDART00000138602
|
stxbp5l
|
syntaxin binding protein 5-like |
| chr15_-_2973368 | 0.67 |
ENSDART00000163131
|
zgc:153184
|
zgc:153184 |
| chr12_+_17754859 | 0.66 |
ENSDART00000112119
|
bhlha15
|
basic helix-loop-helix family, member a15 |
| chr17_+_48164536 | 0.66 |
ENSDART00000161750
ENSDART00000156923 |
plekhd1
|
pleckstrin homology domain containing, family D (with coiled-coil domains) member 1 |
| chr9_+_38074082 | 0.65 |
ENSDART00000017833
|
cacnb4a
|
calcium channel, voltage-dependent, beta 4a subunit |
| chr5_-_57655092 | 0.65 |
ENSDART00000074290
|
mia
|
melanoma inhibitory activity |
| chr16_-_26132122 | 0.65 |
ENSDART00000157787
|
lipeb
|
lipase, hormone-sensitive b |
| chr19_+_40248697 | 0.65 |
ENSDART00000151269
|
cdk6
|
cyclin-dependent kinase 6 |
| chr8_-_51753604 | 0.64 |
ENSDART00000007090
|
tbx16
|
T-box 16 |
| chr8_+_53051701 | 0.62 |
ENSDART00000131514
|
nadka
|
NAD kinase a |
| chr19_-_31402429 | 0.62 |
ENSDART00000137292
|
tmem106bb
|
transmembrane protein 106Bb |
| chr8_+_10869183 | 0.62 |
ENSDART00000188111
|
brpf3b
|
bromodomain and PHD finger containing, 3b |
| chr19_+_32321797 | 0.61 |
ENSDART00000167664
|
atxn1a
|
ataxin 1a |
| chr12_-_45304971 | 0.61 |
ENSDART00000186537
ENSDART00000126405 |
fdxr
|
ferredoxin reductase |
| chr22_+_35516440 | 0.61 |
ENSDART00000184191
|
hykk.2
|
hydroxylysine kinase, tandem duplicate 2 |
| chr23_+_25172976 | 0.60 |
ENSDART00000140789
|
si:dkey-151g10.3
|
si:dkey-151g10.3 |
| chr15_-_43873005 | 0.58 |
ENSDART00000190326
|
NOX4
|
NADPH oxidase 4 |
| chr5_-_12063381 | 0.57 |
ENSDART00000026749
|
nipsnap1
|
nipsnap homolog 1 (C. elegans) |
| chr2_-_55903520 | 0.57 |
ENSDART00000128828
|
cplx4c
|
complexin 4c |
| chr6_-_35446110 | 0.57 |
ENSDART00000058773
|
rgs16
|
regulator of G protein signaling 16 |
| chr12_+_18458502 | 0.57 |
ENSDART00000108745
|
rnf151
|
ring finger protein 151 |
| chr20_-_33675676 | 0.56 |
ENSDART00000147168
|
rock2b
|
rho-associated, coiled-coil containing protein kinase 2b |
| chr14_+_26759332 | 0.56 |
ENSDART00000088484
|
ahnak
|
AHNAK nucleoprotein |
| chr10_+_34321492 | 0.55 |
ENSDART00000190523
|
stard13a
|
StAR-related lipid transfer (START) domain containing 13a |
| chr9_-_55586151 | 0.54 |
ENSDART00000181886
|
arsh
|
arylsulfatase H |
| chr21_+_5993188 | 0.54 |
ENSDART00000048399
|
slc4a4b
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4b |
| chr6_-_46398584 | 0.52 |
ENSDART00000193098
|
camk1a
|
calcium/calmodulin-dependent protein kinase Ia |
| chr15_+_2475894 | 0.51 |
ENSDART00000035939
|
panx1a
|
pannexin 1a |
| chr17_-_51262430 | 0.50 |
ENSDART00000163222
|
trappc12
|
trafficking protein particle complex 12 |
| chr5_-_24124118 | 0.50 |
ENSDART00000051550
|
capga
|
capping protein (actin filament), gelsolin-like a |
| chr2_-_1569250 | 0.50 |
ENSDART00000167202
|
dab1b
|
Dab, reelin signal transducer, homolog 1b (Drosophila) |
| chr3_-_45298487 | 0.49 |
ENSDART00000102245
|
pdpk1a
|
3-phosphoinositide dependent protein kinase 1a |
| chr4_+_54973580 | 0.49 |
ENSDART00000162800
|
si:dkey-56m15.5
|
si:dkey-56m15.5 |
| chr20_-_28698172 | 0.49 |
ENSDART00000190635
|
sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chr4_+_54583639 | 0.49 |
ENSDART00000190527
|
si:ch211-227e10.1
|
si:ch211-227e10.1 |
| chr5_-_66702479 | 0.49 |
ENSDART00000129197
|
mn1b
|
meningioma 1b |
| chr25_+_10416583 | 0.48 |
ENSDART00000073907
|
ehf
|
ets homologous factor |
| chr3_-_34753605 | 0.46 |
ENSDART00000000160
|
thraa
|
thyroid hormone receptor alpha a |
| chr25_-_11016675 | 0.45 |
ENSDART00000099572
|
mespab
|
mesoderm posterior ab |
| chr18_-_33080454 | 0.45 |
ENSDART00000191907
|
v2ra18
|
vomeronasal 2 receptor, a18 |
| chr5_+_30624183 | 0.45 |
ENSDART00000141444
|
abcg4a
|
ATP-binding cassette, sub-family G (WHITE), member 4a |
| chr2_+_28453338 | 0.44 |
ENSDART00000020456
|
mmp15b
|
matrix metallopeptidase 15b |
| chr15_+_46606090 | 0.44 |
ENSDART00000020921
|
CABZ01044048.1
|
|
| chr19_+_9186175 | 0.44 |
ENSDART00000039325
|
hcn3
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 3 |
| chr15_+_5973909 | 0.42 |
ENSDART00000126886
ENSDART00000189618 |
igsf5b
|
immunoglobulin superfamily, member 5b |
| chr24_+_19863246 | 0.42 |
ENSDART00000165242
|
CU638714.1
|
|
| chr4_-_30349370 | 0.42 |
ENSDART00000161790
|
znf1047
|
zinc finger protein 1047 |
| chr5_+_26765275 | 0.41 |
ENSDART00000144169
|
si:ch211-102c2.8
|
si:ch211-102c2.8 |
| chr22_-_34551568 | 0.41 |
ENSDART00000148147
|
rnf123
|
ring finger protein 123 |
| chr9_-_53666031 | 0.41 |
ENSDART00000126314
|
pcdh8
|
protocadherin 8 |
| chr25_+_33033633 | 0.41 |
ENSDART00000192336
|
tln2b
|
talin 2b |
| chr11_-_8782871 | 0.41 |
ENSDART00000158546
|
si:ch211-51h4.2
|
si:ch211-51h4.2 |
| chr5_-_29122834 | 0.39 |
ENSDART00000087197
|
whrnb
|
whirlin b |
| chr4_-_14642379 | 0.39 |
ENSDART00000114977
|
si:ch211-127b11.1
|
si:ch211-127b11.1 |
| chr14_+_21722235 | 0.38 |
ENSDART00000183667
|
stx3a
|
syntaxin 3A |
| chr22_-_16755885 | 0.38 |
ENSDART00000036467
|
patj
|
PATJ, crumbs cell polarity complex component |
| chr9_-_1604601 | 0.37 |
ENSDART00000143130
|
agps
|
alkylglycerone phosphate synthase |
| chr19_+_30990129 | 0.37 |
ENSDART00000052169
ENSDART00000193376 |
sync
|
syncoilin, intermediate filament protein |
| chr25_+_2361721 | 0.37 |
ENSDART00000172905
|
zmp:0000000932
|
zmp:0000000932 |
| chr1_-_11519934 | 0.37 |
ENSDART00000162060
|
sdk1b
|
sidekick cell adhesion molecule 1b |
| chr8_-_8446430 | 0.36 |
ENSDART00000137382
|
cdk16
|
cyclin-dependent kinase 16 |
| chr22_-_36750589 | 0.36 |
ENSDART00000010824
|
acy1
|
aminoacylase 1 |
| chr7_-_37812176 | 0.36 |
ENSDART00000164485
|
adcy7
|
adenylate cyclase 7 |
| chr19_+_42469058 | 0.36 |
ENSDART00000076915
|
si:dkey-166k12.1
|
si:dkey-166k12.1 |
| chr1_-_35113974 | 0.35 |
ENSDART00000192811
ENSDART00000167461 |
BX897691.1
|
|
| chr4_-_20521441 | 0.35 |
ENSDART00000066895
|
rassf8b
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8b |
| chr10_-_39052264 | 0.35 |
ENSDART00000144036
|
igsf5a
|
immunoglobulin superfamily, member 5a |
| chr17_-_7440397 | 0.35 |
ENSDART00000162597
|
grm1b
|
glutamate receptor, metabotropic 1b |
| chr23_-_26227805 | 0.32 |
ENSDART00000158082
|
BX927204.1
|
|
| chr6_+_515181 | 0.31 |
ENSDART00000171374
|
CACNA1I (1 of many)
|
si:ch73-379f7.5 |
| chr3_+_11926030 | 0.31 |
ENSDART00000081367
|
dnaja3a
|
DnaJ (Hsp40) homolog, subfamily A, member 3A |
| chr22_+_3914318 | 0.30 |
ENSDART00000188774
ENSDART00000082034 |
FO904903.1
|
Danio rerio major histocompatibility complex class I ULA (mhc1ula), mRNA. |
| chr4_+_11375894 | 0.30 |
ENSDART00000190471
ENSDART00000143963 |
pcloa
|
piccolo presynaptic cytomatrix protein a |
| chr23_-_46040618 | 0.30 |
ENSDART00000161415
|
CABZ01080918.1
|
|
| chr21_+_34814444 | 0.29 |
ENSDART00000161816
|
wdr55
|
WD repeat domain 55 |
| chr1_+_55643198 | 0.28 |
ENSDART00000060693
|
adgre7
|
adhesion G protein-coupled receptor E7 |
| chr7_-_8022741 | 0.28 |
ENSDART00000172841
|
si:ch211-163c2.1
|
si:ch211-163c2.1 |
| chr15_-_40041455 | 0.28 |
ENSDART00000099359
|
lpar5b
|
lysophosphatidic acid receptor 5b |
| chr1_-_46875493 | 0.28 |
ENSDART00000115081
|
agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr21_+_39948300 | 0.27 |
ENSDART00000137740
|
slc47a1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr19_+_791538 | 0.27 |
ENSDART00000146554
ENSDART00000138406 |
tmem79a
|
transmembrane protein 79a |
| chr21_-_14966718 | 0.27 |
ENSDART00000151200
|
mmp17a
|
matrix metallopeptidase 17a |
| chr15_+_47492838 | 0.26 |
ENSDART00000122372
|
sik3
|
SIK family kinase 3 |
| chr6_-_59381391 | 0.26 |
ENSDART00000157066
|
ppp1r16b
|
protein phosphatase 1, regulatory subunit 16B |
| chr11_+_42556395 | 0.26 |
ENSDART00000039206
|
rps23
|
ribosomal protein S23 |
| chr3_-_60886984 | 0.26 |
ENSDART00000170974
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr23_-_37432459 | 0.25 |
ENSDART00000188916
|
zmp:0000001088
|
zmp:0000001088 |
| chr7_+_36467796 | 0.25 |
ENSDART00000146202
|
aktip
|
akt interacting protein |
| chr10_-_38456382 | 0.24 |
ENSDART00000182129
|
gdpd5a
|
glycerophosphodiester phosphodiesterase domain containing 5a |
| chr15_-_42736433 | 0.24 |
ENSDART00000154379
|
si:ch211-181d7.1
|
si:ch211-181d7.1 |
| chr24_-_21404367 | 0.24 |
ENSDART00000152093
|
atp8a2
|
ATPase phospholipid transporting 8A2 |
| chr7_-_52842605 | 0.24 |
ENSDART00000083002
|
map1aa
|
microtubule-associated protein 1Aa |
| chr16_+_46725087 | 0.24 |
ENSDART00000008920
|
rab11al
|
RAB11a, member RAS oncogene family, like |
| chr22_+_38159823 | 0.23 |
ENSDART00000104527
|
tm4sf18
|
transmembrane 4 L six family member 18 |
| chr5_-_29122615 | 0.23 |
ENSDART00000144802
|
whrnb
|
whirlin b |
| chr20_+_23498255 | 0.23 |
ENSDART00000149922
|
palld
|
palladin, cytoskeletal associated protein |
| chr2_+_30463825 | 0.23 |
ENSDART00000092356
|
neto1
|
neuropilin (NRP) and tolloid (TLL)-like 1 |
| chr22_-_36770649 | 0.23 |
ENSDART00000169442
|
acy1
|
aminoacylase 1 |
| chr12_+_2381213 | 0.23 |
ENSDART00000188007
|
LO018238.1
|
|
| chr23_-_21946603 | 0.23 |
ENSDART00000148120
ENSDART00000184524 |
ephb2a
|
eph receptor B2a |
| chr4_-_75982054 | 0.23 |
ENSDART00000168369
|
si:ch211-232d10.1
|
si:ch211-232d10.1 |
| chr15_-_12319065 | 0.23 |
ENSDART00000162973
ENSDART00000170543 |
fxyd6
|
FXYD domain containing ion transport regulator 6 |
| chr19_-_9867001 | 0.22 |
ENSDART00000091695
|
cacng7a
|
calcium channel, voltage-dependent, gamma subunit 7a |
| chr25_-_19666107 | 0.22 |
ENSDART00000149889
|
atp2b1b
|
ATPase plasma membrane Ca2+ transporting 1b |
| chr23_-_39959784 | 0.22 |
ENSDART00000115046
|
xcr1a.1
|
chemokine (C motif) receptor 1a, duplicate 1 |
| chr12_-_44016898 | 0.22 |
ENSDART00000175304
|
si:dkey-201i2.4
|
si:dkey-201i2.4 |
| chr23_-_29812667 | 0.22 |
ENSDART00000006120
|
pik3cd
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta |
| chr8_-_18229169 | 0.22 |
ENSDART00000131764
ENSDART00000143036 ENSDART00000145986 |
si:ch211-241d21.5
|
si:ch211-241d21.5 |
| chr14_-_31814149 | 0.21 |
ENSDART00000173393
|
arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr2_-_57918314 | 0.21 |
ENSDART00000138265
|
si:dkeyp-68b7.7
|
si:dkeyp-68b7.7 |
| chr19_-_29302249 | 0.21 |
ENSDART00000188751
|
srfbp1
|
serum response factor binding protein 1 |
| chr5_+_57726425 | 0.21 |
ENSDART00000134684
|
fdxacb1
|
ferredoxin-fold anticodon binding domain containing 1 |
| chr24_-_11057305 | 0.21 |
ENSDART00000186494
|
asap1b
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1b |
| chr17_-_22324727 | 0.21 |
ENSDART00000160341
|
CU104709.1
|
|
| chr6_+_13606410 | 0.20 |
ENSDART00000104716
|
asic4b
|
acid-sensing (proton-gated) ion channel family member 4b |
| chr1_+_56779495 | 0.20 |
ENSDART00000192871
|
FO680692.1
|
|
| chr16_+_27444098 | 0.20 |
ENSDART00000157690
|
invs
|
inversin |
| chr7_-_35083585 | 0.20 |
ENSDART00000192732
|
agrp
|
agouti related neuropeptide |
| chr14_-_36763302 | 0.19 |
ENSDART00000074786
|
ctso
|
cathepsin O |
| chr22_+_16022211 | 0.19 |
ENSDART00000062618
|
serpinc1
|
serpin peptidase inhibitor, clade C (antithrombin), member 1 |
| chr5_-_23855447 | 0.19 |
ENSDART00000051541
|
gbgt1l3
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 3 |
| chr3_-_32603191 | 0.19 |
ENSDART00000150997
|
si:ch73-248e21.7
|
si:ch73-248e21.7 |
| chr9_+_51655636 | 0.19 |
ENSDART00000169908
|
RBMS1 (1 of many)
|
RNA binding motif single stranded interacting protein 1 |
| chr8_-_54223316 | 0.18 |
ENSDART00000018054
|
trh
|
thyrotropin-releasing hormone |
| chr11_+_24620742 | 0.18 |
ENSDART00000182471
ENSDART00000048365 |
syt6b
|
synaptotagmin VIb |
| chr5_-_38248347 | 0.18 |
ENSDART00000084917
ENSDART00000139479 |
slc12a9
|
solute carrier family 12, member 9 |
| chr19_+_42470396 | 0.18 |
ENSDART00000191679
|
si:dkey-166k12.1
|
si:dkey-166k12.1 |
| chr16_-_14332561 | 0.18 |
ENSDART00000186784
ENSDART00000011224 |
itga10
|
integrin, alpha 10 |
| chr23_-_17003533 | 0.18 |
ENSDART00000080545
|
dnmt3bb.2
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate b.2 |
| chr16_+_20934353 | 0.18 |
ENSDART00000052660
|
skap2
|
src kinase associated phosphoprotein 2 |
| chr18_+_13182528 | 0.16 |
ENSDART00000166298
|
zgc:56622
|
zgc:56622 |
| chr5_+_5689476 | 0.16 |
ENSDART00000022729
|
unm_sa808
|
un-named sa808 |
| chr21_+_18877130 | 0.16 |
ENSDART00000136893
|
si:dkey-65l23.2
|
si:dkey-65l23.2 |
| chr14_+_25817628 | 0.16 |
ENSDART00000047680
|
glra1
|
glycine receptor, alpha 1 |
| chr18_+_2593756 | 0.16 |
ENSDART00000158022
|
p2ry2.3
|
purinergic receptor P2Y2, tandem duplicate 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx3.3-1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0033512 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.4 | 2.9 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.3 | 2.1 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.3 | 0.9 | GO:1900274 | positive regulation of phospholipase C activity(GO:0010863) regulation of phospholipase C activity(GO:1900274) |
| 0.2 | 1.0 | GO:0019068 | virion assembly(GO:0019068) virus maturation(GO:0019075) multi-organism membrane organization(GO:0044803) viral budding(GO:0046755) multi-organism organelle organization(GO:1902590) multi-organism membrane budding(GO:1902592) |
| 0.2 | 2.6 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.2 | 1.3 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.2 | 0.9 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.2 | 0.6 | GO:0048389 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) intermediate mesoderm development(GO:0048389) |
| 0.2 | 1.0 | GO:0060251 | regulation of glial cell proliferation(GO:0060251) |
| 0.2 | 0.8 | GO:0039689 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.1 | 0.8 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 1.5 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 0.4 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.1 | 1.0 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.1 | 0.8 | GO:0000050 | urea cycle(GO:0000050) urea metabolic process(GO:0019627) |
| 0.1 | 2.1 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.7 | GO:0006833 | water transport(GO:0006833) |
| 0.1 | 0.6 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 1.4 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.1 | 0.8 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 0.4 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.1 | 0.5 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.1 | 0.6 | GO:0043696 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.1 | 0.2 | GO:2000257 | regulation of protein activation cascade(GO:2000257) |
| 0.1 | 0.5 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.1 | 0.2 | GO:1903792 | histamine metabolic process(GO:0001692) negative regulation of anion transport(GO:1903792) |
| 0.1 | 0.8 | GO:1901642 | nucleoside transport(GO:0015858) nucleoside transmembrane transport(GO:1901642) |
| 0.1 | 0.8 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 0.2 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.1 | 0.2 | GO:0072003 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.0 | 1.5 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.5 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.0 | 1.1 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.6 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.4 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.3 | GO:0060465 | pharynx development(GO:0060465) |
| 0.0 | 0.4 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.2 | GO:0070257 | regulation of mucus secretion(GO:0070255) positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.4 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.6 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.6 | GO:2000651 | positive regulation of sodium ion transport(GO:0010765) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 1.6 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.6 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.3 | GO:0035188 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.2 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.0 | 0.1 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) Schwann cell migration(GO:0036135) cardiac neuron differentiation(GO:0060945) cardiac neuron development(GO:0060959) |
| 0.0 | 0.6 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.6 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.3 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.0 | 0.5 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.2 | GO:2000253 | adult feeding behavior(GO:0008343) positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.8 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.2 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.0 | 0.2 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.8 | GO:0061035 | regulation of cartilage development(GO:0061035) |
| 0.0 | 0.2 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.0 | 0.7 | GO:0050708 | regulation of protein secretion(GO:0050708) |
| 0.0 | 0.1 | GO:0032096 | negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.0 | 0.7 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.5 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.0 | 0.2 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 0.2 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.1 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.0 | 0.3 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 1.0 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.0 | 1.2 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.2 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.7 | GO:0031017 | exocrine pancreas development(GO:0031017) exocrine system development(GO:0035272) |
| 0.0 | 0.7 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.2 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.3 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.1 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.1 | 1.5 | GO:0098894 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.1 | 0.6 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 1.1 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 0.6 | GO:0032426 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) stereocilium tip(GO:0032426) |
| 0.1 | 1.0 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 2.1 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.8 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.6 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.5 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.3 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.0 | 0.8 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.6 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 1.6 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.5 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 2.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.4 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 1.4 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.6 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.6 | GO:0099634 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.7 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.2 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.3 | 1.4 | GO:2001070 | starch binding(GO:2001070) |
| 0.2 | 2.6 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.1 | 1.5 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 1.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.6 | GO:0019202 | amino acid kinase activity(GO:0019202) |
| 0.1 | 0.5 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.1 | 2.1 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 0.8 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.7 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.1 | 1.0 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 1.0 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.1 | 0.9 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.6 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.1 | 0.2 | GO:0016436 | rRNA (uridine) methyltransferase activity(GO:0016436) |
| 0.1 | 1.0 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.8 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.3 | GO:0017020 | myosin phosphatase regulator activity(GO:0017020) |
| 0.1 | 0.6 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 1.5 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.1 | 0.8 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.1 | 0.2 | GO:0031781 | type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.3 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.0 | 1.4 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.6 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.6 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.7 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.6 | GO:0019870 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 1.1 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.2 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.2 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.5 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.3 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.2 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.3 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.0 | 1.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.2 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.6 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.3 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.4 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.4 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.0 | 0.5 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.6 | GO:0015370 | solute:sodium symporter activity(GO:0015370) |
| 0.0 | 0.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.2 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 0.2 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 1.1 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.8 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.0 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.6 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.6 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 1.0 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.5 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 1.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.9 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.4 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.8 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.6 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 2.3 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.2 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |