Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
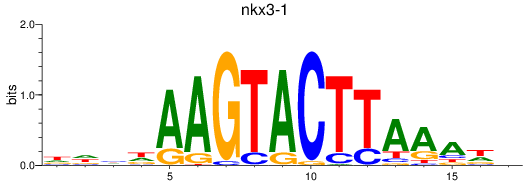
Results for nkx3-1
Z-value: 0.55
Transcription factors associated with nkx3-1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx3-1
|
ENSDARG00000078280 | NK3 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nkx3-1 | dr11_v1_chr8_-_50259448_50259448 | -0.42 | 7.1e-02 | Click! |
Activity profile of nkx3-1 motif
Sorted Z-values of nkx3-1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_22680560 | 0.99 |
ENSDART00000133761
|
ponzr4
|
plac8 onzin related protein 4 |
| chr9_-_22158325 | 0.97 |
ENSDART00000114568
|
crygm2d16
|
crystallin, gamma M2d16 |
| chr9_-_22158784 | 0.95 |
ENSDART00000167850
|
crygm2d14
|
crystallin, gamma M2d14 |
| chr14_-_33894915 | 0.92 |
ENSDART00000143290
|
urp1
|
urotensin-related peptide 1 |
| chr1_-_26293203 | 0.85 |
ENSDART00000180140
|
cxxc4
|
CXXC finger 4 |
| chr21_+_10828828 | 0.79 |
ENSDART00000136449
|
oacyl
|
O-acyltransferase like |
| chr5_-_64883082 | 0.76 |
ENSDART00000064983
ENSDART00000139066 |
krt1-c5
|
keratin, type 1, gene c5 |
| chr11_-_30636163 | 0.65 |
ENSDART00000140516
|
zgc:153665
|
zgc:153665 |
| chr22_-_11626014 | 0.62 |
ENSDART00000063133
ENSDART00000160085 |
gcga
|
glucagon a |
| chr5_+_38276582 | 0.59 |
ENSDART00000158532
|
gnb2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr19_-_42556086 | 0.56 |
ENSDART00000051731
|
si:dkey-267n13.1
|
si:dkey-267n13.1 |
| chr12_-_16595406 | 0.56 |
ENSDART00000166798
|
si:dkey-239j18.2
|
si:dkey-239j18.2 |
| chr12_-_16595177 | 0.55 |
ENSDART00000133962
|
si:dkey-239j18.2
|
si:dkey-239j18.2 |
| chr23_+_35672542 | 0.53 |
ENSDART00000046268
|
pmelb
|
premelanosome protein b |
| chr11_-_24191928 | 0.53 |
ENSDART00000136827
|
sox12
|
SRY (sex determining region Y)-box 12 |
| chr1_-_26292897 | 0.51 |
ENSDART00000112899
ENSDART00000185410 |
cxxc4
|
CXXC finger 4 |
| chr11_-_36350421 | 0.51 |
ENSDART00000141477
|
psma5
|
proteasome subunit alpha 5 |
| chr19_+_41419519 | 0.51 |
ENSDART00000148732
ENSDART00000149799 |
dlx6a
|
distal-less homeobox 6a |
| chr25_-_13703826 | 0.49 |
ENSDART00000163398
|
pla2g15
|
phospholipase A2, group XV |
| chr22_-_10541712 | 0.48 |
ENSDART00000013933
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr6_-_19351495 | 0.48 |
ENSDART00000164287
|
grb2a
|
growth factor receptor-bound protein 2a |
| chr12_+_14079097 | 0.45 |
ENSDART00000078033
|
si:ch211-217a12.1
|
si:ch211-217a12.1 |
| chr18_-_16801033 | 0.45 |
ENSDART00000100100
|
admb
|
adrenomedullin b |
| chr7_-_48667056 | 0.44 |
ENSDART00000006378
|
cdkn1ca
|
cyclin-dependent kinase inhibitor 1Ca |
| chr4_+_76705830 | 0.43 |
ENSDART00000064312
|
ms4a17a.7
|
membrane-spanning 4-domains, subfamily A, member 17A.7 |
| chr11_-_7156620 | 0.43 |
ENSDART00000172823
ENSDART00000172879 ENSDART00000078916 |
smim7
|
small integral membrane protein 7 |
| chr5_+_51594209 | 0.43 |
ENSDART00000164668
ENSDART00000058403 ENSDART00000055857 |
ckmt2b
|
creatine kinase, mitochondrial 2b (sarcomeric) |
| chr17_-_19344999 | 0.42 |
ENSDART00000138315
|
gsc
|
goosecoid |
| chr22_-_11623063 | 0.41 |
ENSDART00000145653
|
gcga
|
glucagon a |
| chr8_-_40464935 | 0.41 |
ENSDART00000040013
|
myl7
|
myosin, light chain 7, regulatory |
| chr15_-_24676211 | 0.40 |
ENSDART00000153502
ENSDART00000100756 |
tmem199
|
transmembrane protein 199 |
| chr9_+_35016201 | 0.40 |
ENSDART00000182404
|
gabpa
|
GA binding protein transcription factor, alpha subunit |
| chr12_-_10476448 | 0.39 |
ENSDART00000106172
|
rac1a
|
Rac family small GTPase 1a |
| chr24_-_26328721 | 0.38 |
ENSDART00000125468
|
apodb
|
apolipoprotein Db |
| chr25_+_14017609 | 0.38 |
ENSDART00000129105
ENSDART00000125733 |
chst1
|
carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 |
| chr2_+_7295515 | 0.38 |
ENSDART00000152987
|
si:dkeyp-106c3.1
|
si:dkeyp-106c3.1 |
| chr17_+_8925232 | 0.38 |
ENSDART00000036668
|
psmc1a
|
proteasome 26S subunit, ATPase 1a |
| chr22_-_12763350 | 0.38 |
ENSDART00000143574
|
cnot9
|
CCR4-NOT transcription complex subunit 9 |
| chr13_-_32726178 | 0.36 |
ENSDART00000012232
|
pdss2
|
prenyl (decaprenyl) diphosphate synthase, subunit 2 |
| chr2_-_38000276 | 0.36 |
ENSDART00000034790
|
pcp4l1
|
Purkinje cell protein 4 like 1 |
| chr10_+_21588002 | 0.35 |
ENSDART00000100596
|
pcdh1a6
|
protocadherin 1 alpha 6 |
| chr4_-_72609735 | 0.35 |
ENSDART00000174299
ENSDART00000159227 |
si:cabz01054394.6
|
si:cabz01054394.6 |
| chr18_-_14691727 | 0.35 |
ENSDART00000010129
|
pdf
|
peptide deformylase, mitochondrial |
| chr23_-_40295610 | 0.35 |
ENSDART00000186719
|
CABZ01067002.1
|
|
| chr17_-_43623356 | 0.34 |
ENSDART00000075596
ENSDART00000136431 |
rtkn2a
|
rhotekin 2a |
| chr19_-_1405363 | 0.33 |
ENSDART00000165516
|
cfap20
|
cilia and flagella associated protein 20 |
| chr19_+_20793237 | 0.32 |
ENSDART00000014774
|
txnl4a
|
thioredoxin-like 4A |
| chr3_-_15999501 | 0.31 |
ENSDART00000160668
|
nme3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr1_-_55248496 | 0.31 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr15_+_2534740 | 0.31 |
ENSDART00000138469
|
cux1b
|
cut-like homeobox 1b |
| chr5_-_37900350 | 0.30 |
ENSDART00000084839
ENSDART00000084841 ENSDART00000133437 |
tmprss13b
|
transmembrane protease, serine 13b |
| chr4_+_17279966 | 0.30 |
ENSDART00000067005
ENSDART00000137487 |
bcat1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr19_-_3821678 | 0.30 |
ENSDART00000169639
|
si:dkey-206d17.12
|
si:dkey-206d17.12 |
| chr20_-_33507458 | 0.30 |
ENSDART00000140287
|
paplnb
|
papilin b, proteoglycan-like sulfated glycoprotein |
| chr7_+_37742299 | 0.30 |
ENSDART00000143300
|
brd7
|
bromodomain containing 7 |
| chr10_+_20128267 | 0.29 |
ENSDART00000064615
|
dmtn
|
dematin actin binding protein |
| chr8_+_4761244 | 0.29 |
ENSDART00000137252
ENSDART00000132139 ENSDART00000034968 ENSDART00000143139 |
ufd1l
|
ubiquitin recognition factor in ER associated degradation 1 |
| chr4_+_842010 | 0.29 |
ENSDART00000067461
|
si:ch211-152c2.3
|
si:ch211-152c2.3 |
| chr4_-_17805128 | 0.29 |
ENSDART00000128988
|
spi2
|
Spi-2 proto-oncogene |
| chr12_-_44070043 | 0.29 |
ENSDART00000180059
|
si:ch73-329n5.3
|
si:ch73-329n5.3 |
| chr13_+_28903959 | 0.28 |
ENSDART00000166435
|
si:ch73-28h20.1
|
si:ch73-28h20.1 |
| chr24_-_8261786 | 0.28 |
ENSDART00000106388
|
eef1e1
|
eukaryotic translation elongation factor 1 epsilon 1 |
| chr24_+_16140423 | 0.28 |
ENSDART00000105955
|
TIMM21
|
si:dkey-118j18.1 |
| chr1_+_57040472 | 0.28 |
ENSDART00000181365
|
si:ch211-1f22.16
|
si:ch211-1f22.16 |
| chr7_-_34192834 | 0.28 |
ENSDART00000125131
|
smad6a
|
SMAD family member 6a |
| chr2_+_15100742 | 0.28 |
ENSDART00000027171
|
f3b
|
coagulation factor IIIb |
| chr8_-_34065573 | 0.27 |
ENSDART00000186946
|
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr17_+_24615091 | 0.27 |
ENSDART00000064739
|
rpl13a
|
ribosomal protein L13a |
| chr2_-_11101329 | 0.27 |
ENSDART00000164416
|
lrrc53
|
leucine rich repeat containing 53 |
| chr4_-_72544805 | 0.26 |
ENSDART00000110261
|
zgc:175107
|
zgc:175107 |
| chr6_+_21993613 | 0.26 |
ENSDART00000160750
ENSDART00000083070 |
thumpd3
|
THUMP domain containing 3 |
| chr22_-_5252005 | 0.26 |
ENSDART00000132942
ENSDART00000081801 |
ncln
|
nicalin |
| chr1_-_38813679 | 0.26 |
ENSDART00000148917
|
asb5b
|
ankyrin repeat and SOCS box containing 5b |
| chr3_-_20957011 | 0.25 |
ENSDART00000159787
ENSDART00000180531 |
ndufa4l
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 4, like |
| chr16_-_41465542 | 0.25 |
ENSDART00000169116
ENSDART00000187446 |
cpne4a
|
copine IVa |
| chr1_+_580642 | 0.25 |
ENSDART00000147633
|
mrpl39
|
mitochondrial ribosomal protein L39 |
| chr6_-_54826061 | 0.25 |
ENSDART00000149982
|
tnni1b
|
troponin I type 1b (skeletal, slow) |
| chr4_+_76735113 | 0.25 |
ENSDART00000075602
|
ms4a17a.6
|
membrane-spanning 4-domains, subfamily A, member 17A.6 |
| chr19_-_10330778 | 0.25 |
ENSDART00000081465
ENSDART00000136653 ENSDART00000171232 |
ccdc106b
|
coiled-coil domain containing 106b |
| chr25_-_24240797 | 0.25 |
ENSDART00000132790
|
spty2d1
|
SPT2 chromatin protein domain containing 1 |
| chr14_+_48903640 | 0.25 |
ENSDART00000165323
|
zgc:154054
|
zgc:154054 |
| chr21_-_25623342 | 0.24 |
ENSDART00000101201
|
tmem223
|
transmembrane protein 223 |
| chr1_-_49950643 | 0.24 |
ENSDART00000138301
|
sgms2
|
sphingomyelin synthase 2 |
| chr12_-_25150239 | 0.24 |
ENSDART00000038415
ENSDART00000135368 |
rhoq
|
ras homolog family member Q |
| chr15_-_4415917 | 0.24 |
ENSDART00000062874
|
atp1b3b
|
ATPase Na+/K+ transporting subunit beta 3b |
| chr12_-_33579873 | 0.24 |
ENSDART00000184661
|
tdrkh
|
tudor and KH domain containing |
| chr5_-_52752514 | 0.24 |
ENSDART00000190162
|
fam189a2
|
family with sequence similarity 189, member A2 |
| chr14_-_33978117 | 0.24 |
ENSDART00000128515
|
foxa
|
forkhead box A sequence |
| chr10_-_44924289 | 0.24 |
ENSDART00000171267
|
tuba7l
|
tubulin, alpha 7 like |
| chr12_+_28986308 | 0.23 |
ENSDART00000153178
|
si:rp71-1c10.8
|
si:rp71-1c10.8 |
| chr15_+_36309070 | 0.23 |
ENSDART00000157034
|
gmnc
|
geminin coiled-coil domain containing |
| chr19_-_3916075 | 0.23 |
ENSDART00000161799
|
si:ch73-281i18.7
|
si:ch73-281i18.7 |
| chr19_+_7424347 | 0.23 |
ENSDART00000004622
|
sf3b4
|
splicing factor 3b, subunit 4 |
| chr3_-_16000168 | 0.22 |
ENSDART00000160628
|
nme3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr20_-_33497128 | 0.22 |
ENSDART00000101974
|
erh
|
enhancer of rudimentary homolog (Drosophila) |
| chr24_+_14595680 | 0.22 |
ENSDART00000137337
ENSDART00000091784 ENSDART00000136026 |
thtpa
|
thiamine triphosphatase |
| chr5_+_26795773 | 0.22 |
ENSDART00000145631
|
tcn2
|
transcobalamin II |
| chr16_+_31922065 | 0.22 |
ENSDART00000131661
ENSDART00000144194 ENSDART00000145510 |
rps9
|
ribosomal protein S9 |
| chr15_+_41836104 | 0.22 |
ENSDART00000137434
|
pxylp1
|
2-phosphoxylose phosphatase 1 |
| chr22_+_12770877 | 0.22 |
ENSDART00000044683
|
ftcd
|
formimidoyltransferase cyclodeaminase |
| chr17_+_24623561 | 0.21 |
ENSDART00000156734
|
thumpd2
|
THUMP domain containing 2 |
| chr3_-_13599482 | 0.21 |
ENSDART00000166639
|
tufm
|
Tu translation elongation factor, mitochondrial |
| chr22_+_24220937 | 0.21 |
ENSDART00000165380
|
uchl5
|
ubiquitin carboxyl-terminal hydrolase L5 |
| chr16_+_33142734 | 0.21 |
ENSDART00000138244
|
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr7_+_31132588 | 0.21 |
ENSDART00000173702
|
tjp1a
|
tight junction protein 1a |
| chr18_+_402048 | 0.21 |
ENSDART00000166345
|
gpib
|
glucose-6-phosphate isomerase b |
| chr2_-_10877228 | 0.21 |
ENSDART00000138718
ENSDART00000034246 |
cdc7
|
cell division cycle 7 homolog (S. cerevisiae) |
| chr15_+_17848590 | 0.21 |
ENSDART00000168940
|
zgc:113279
|
zgc:113279 |
| chr11_+_24900123 | 0.21 |
ENSDART00000044987
ENSDART00000148023 |
timm17a
|
translocase of inner mitochondrial membrane 17 homolog A (yeast) |
| chr7_+_22981441 | 0.21 |
ENSDART00000182887
|
ccnb3
|
cyclin B3 |
| chr24_-_27473771 | 0.21 |
ENSDART00000139874
|
cxl34b.11
|
CX chemokine ligand 34b, duplicate 11 |
| chr4_+_30454496 | 0.20 |
ENSDART00000164555
|
si:dkey-199m13.4
|
si:dkey-199m13.4 |
| chr15_-_39969988 | 0.20 |
ENSDART00000146054
|
rps5
|
ribosomal protein S5 |
| chr6_+_37894914 | 0.20 |
ENSDART00000148817
|
oca2
|
oculocutaneous albinism II |
| chr10_-_34915886 | 0.20 |
ENSDART00000141201
ENSDART00000002166 |
ccna1
|
cyclin A1 |
| chr2_+_37750016 | 0.20 |
ENSDART00000154726
|
si:dkeyp-66d1.7
|
si:dkeyp-66d1.7 |
| chr4_+_17280868 | 0.20 |
ENSDART00000145349
|
bcat1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr20_+_36623807 | 0.20 |
ENSDART00000149171
ENSDART00000062895 |
srp9
|
signal recognition particle 9 |
| chr1_+_26012668 | 0.20 |
ENSDART00000182428
|
BX323448.1
|
|
| chr9_+_48123224 | 0.19 |
ENSDART00000141610
|
klhl23
|
kelch-like family member 23 |
| chr10_+_15310811 | 0.19 |
ENSDART00000136239
|
kcnv2a
|
potassium channel, subfamily V, member 2a |
| chr20_+_33904258 | 0.19 |
ENSDART00000170930
|
rxrgb
|
retinoid X receptor, gamma b |
| chr25_-_173165 | 0.19 |
ENSDART00000193594
|
CABZ01114053.1
|
|
| chr4_+_76745015 | 0.19 |
ENSDART00000155883
|
ms4a17a.10
|
membrane-spanning 4-domains, subfamily A, member 17A.10 |
| chr5_-_69707787 | 0.19 |
ENSDART00000108820
ENSDART00000149692 |
dguok
|
deoxyguanosine kinase |
| chr9_-_46072805 | 0.18 |
ENSDART00000169682
|
hdac4
|
histone deacetylase 4 |
| chr2_+_10642047 | 0.18 |
ENSDART00000091570
|
fam69aa
|
family with sequence similarity 69, member Aa |
| chr18_-_7382003 | 0.18 |
ENSDART00000101238
ENSDART00000178284 |
bbs10
|
Bardet-Biedl syndrome 10 |
| chr20_-_38758797 | 0.18 |
ENSDART00000061394
|
trim54
|
tripartite motif containing 54 |
| chr6_+_72040 | 0.18 |
ENSDART00000122957
|
crygm4
|
crystallin, gamma M4 |
| chr4_+_7514687 | 0.18 |
ENSDART00000067350
ENSDART00000169998 ENSDART00000162082 |
tnnt2e
|
troponin T2e, cardiac |
| chr25_+_22587306 | 0.17 |
ENSDART00000067479
|
stra6
|
stimulated by retinoic acid 6 |
| chr22_+_13886821 | 0.17 |
ENSDART00000130585
ENSDART00000105711 |
sh3bp4a
|
SH3-domain binding protein 4a |
| chr15_+_11427620 | 0.17 |
ENSDART00000168688
|
ndufc2
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2 |
| chr25_+_3318192 | 0.17 |
ENSDART00000146154
|
slc25a3b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3b |
| chr7_-_26532089 | 0.17 |
ENSDART00000121698
|
senp3b
|
SUMO1/sentrin/SMT3 specific peptidase 3b |
| chr23_+_36340520 | 0.16 |
ENSDART00000011201
|
copz1
|
coatomer protein complex, subunit zeta 1 |
| chr5_+_41143563 | 0.16 |
ENSDART00000011229
|
sub1b
|
SUB1 homolog, transcriptional regulator b |
| chr19_-_2420990 | 0.16 |
ENSDART00000181498
|
TMEM196 (1 of many)
|
transmembrane protein 196 |
| chr15_+_19300230 | 0.16 |
ENSDART00000183363
|
jam3a
|
junctional adhesion molecule 3a |
| chr19_+_33139164 | 0.16 |
ENSDART00000043039
|
fam84b
|
family with sequence similarity 84, member B |
| chr23_-_4019699 | 0.16 |
ENSDART00000159780
|
slc9a8
|
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr17_-_24703778 | 0.16 |
ENSDART00000156061
|
si:ch211-15d5.12
|
si:ch211-15d5.12 |
| chr6_+_11438972 | 0.16 |
ENSDART00000029314
|
col5a2b
|
collagen, type V, alpha 2b |
| chr2_-_53525896 | 0.16 |
ENSDART00000057053
|
im:7138239
|
im:7138239 |
| chr18_-_49318823 | 0.15 |
ENSDART00000098419
|
sb:cb81
|
sb:cb81 |
| chr23_+_19813677 | 0.15 |
ENSDART00000139192
ENSDART00000142308 |
emd
|
emerin (Emery-Dreifuss muscular dystrophy) |
| chr22_+_25236657 | 0.15 |
ENSDART00000138012
|
zgc:172218
|
zgc:172218 |
| chr9_-_12888082 | 0.15 |
ENSDART00000133135
ENSDART00000134415 |
si:ch211-167j6.3
|
si:ch211-167j6.3 |
| chr7_-_30280934 | 0.15 |
ENSDART00000126741
|
serf2
|
small EDRK-rich factor 2 |
| chr19_-_31372896 | 0.15 |
ENSDART00000046609
|
scin
|
scinderin |
| chr21_-_11286483 | 0.15 |
ENSDART00000074845
|
rtkn2b
|
rhotekin 2b |
| chr2_+_1487118 | 0.14 |
ENSDART00000147283
|
c8a
|
complement component 8, alpha polypeptide |
| chr25_-_13050959 | 0.14 |
ENSDART00000169041
|
ccl35.1
|
chemokine (C-C motif) ligand 35, duplicate 1 |
| chr7_+_22657566 | 0.14 |
ENSDART00000141048
|
ponzr5
|
plac8 onzin related protein 5 |
| chr13_+_45488193 | 0.14 |
ENSDART00000188004
|
tmem50a
|
transmembrane protein 50A |
| chr17_+_20205185 | 0.14 |
ENSDART00000167007
|
gnrh3
|
gonadotropin-releasing hormone 3 |
| chr20_-_18535502 | 0.14 |
ENSDART00000049437
|
cdc42bpb
|
CDC42 binding protein kinase beta (DMPK-like) |
| chr17_+_22577472 | 0.14 |
ENSDART00000045099
|
yipf4
|
Yip1 domain family, member 4 |
| chr4_-_65323874 | 0.14 |
ENSDART00000175271
ENSDART00000187913 |
BX571813.1
|
|
| chr5_+_36666715 | 0.13 |
ENSDART00000097686
|
zgc:153990
|
zgc:153990 |
| chr21_-_4250682 | 0.13 |
ENSDART00000099389
|
dnlz
|
DNL-type zinc finger |
| chr10_+_26827113 | 0.13 |
ENSDART00000100301
|
sssca1
|
Sjogren syndrome/scleroderma autoantigen 1 |
| chr11_+_24729346 | 0.13 |
ENSDART00000087740
|
zgc:153953
|
zgc:153953 |
| chr16_-_32008277 | 0.13 |
ENSDART00000102133
ENSDART00000141512 |
gstk4
|
glutathione S-transferase kappa 4 |
| chr15_+_45640906 | 0.13 |
ENSDART00000149361
ENSDART00000149079 |
sagb
|
S-antigen; retina and pineal gland (arrestin) b |
| chr9_+_48123002 | 0.13 |
ENSDART00000099794
ENSDART00000169733 |
klhl23
|
kelch-like family member 23 |
| chr4_-_43280244 | 0.12 |
ENSDART00000150762
|
si:dkeyp-53e4.1
|
si:dkeyp-53e4.1 |
| chr5_+_30596632 | 0.12 |
ENSDART00000051414
|
hinfp
|
histone H4 transcription factor |
| chr4_-_36793658 | 0.12 |
ENSDART00000172152
|
si:dkeyp-87d1.1
|
si:dkeyp-87d1.1 |
| chr9_-_8296723 | 0.12 |
ENSDART00000139867
|
si:ch211-145c1.1
|
si:ch211-145c1.1 |
| chr16_-_47427016 | 0.12 |
ENSDART00000074575
|
sept7b
|
septin 7b |
| chr17_+_30894431 | 0.12 |
ENSDART00000127996
|
degs2
|
delta(4)-desaturase, sphingolipid 2 |
| chr17_-_1705013 | 0.12 |
ENSDART00000182864
|
CABZ01086293.1
|
|
| chr25_+_3788074 | 0.12 |
ENSDART00000154008
|
chid1
|
chitinase domain containing 1 |
| chr1_+_55752593 | 0.12 |
ENSDART00000108838
ENSDART00000134770 |
tecrb
|
trans-2,3-enoyl-CoA reductase b |
| chr5_+_30596477 | 0.12 |
ENSDART00000124487
|
hinfp
|
histone H4 transcription factor |
| chr3_-_18189283 | 0.12 |
ENSDART00000049240
|
tob1a
|
transducer of ERBB2, 1a |
| chr5_-_24997293 | 0.11 |
ENSDART00000177478
|
emid1
|
EMI domain containing 1 |
| chr13_+_36680564 | 0.11 |
ENSDART00000136030
ENSDART00000006923 |
atp5s
|
ATP synthase, H+ transporting, mitochondrial F0 complex subunit s |
| chr7_-_26627252 | 0.11 |
ENSDART00000164050
ENSDART00000159826 |
phf23b
|
PHD finger protein 23b |
| chr11_+_30636351 | 0.11 |
ENSDART00000087909
|
tmem246
|
transmembrane protein 246 |
| chr17_+_6765621 | 0.11 |
ENSDART00000156637
ENSDART00000007622 |
afg1la
|
AFG1 like ATPase a |
| chr7_+_58179513 | 0.11 |
ENSDART00000123117
|
ggh
|
gamma-glutamyl hydrolase (conjugase, folylpolygammaglutamyl hydrolase) |
| chr4_-_52621665 | 0.11 |
ENSDART00000137064
|
si:dkeyp-104f11.6
|
si:dkeyp-104f11.6 |
| chr6_+_10338554 | 0.11 |
ENSDART00000186936
|
cobll1a
|
cordon-bleu WH2 repeat protein-like 1a |
| chr17_-_43031763 | 0.11 |
ENSDART00000132754
ENSDART00000050399 |
npc2
|
Niemann-Pick disease, type C2 |
| chr20_+_25911342 | 0.11 |
ENSDART00000146004
|
ttbk2b
|
tau tubulin kinase 2b |
| chr5_-_10082244 | 0.11 |
ENSDART00000036421
|
chek2
|
checkpoint kinase 2 |
| chr19_-_18152407 | 0.11 |
ENSDART00000193264
ENSDART00000016135 |
nfe2l3
|
nuclear factor, erythroid 2-like 3 |
| chr21_-_27201495 | 0.11 |
ENSDART00000145013
|
zgc:77262
|
zgc:77262 |
| chr20_-_34985555 | 0.10 |
ENSDART00000037065
|
sccpdhb
|
saccharopine dehydrogenase b |
| chr7_+_17947217 | 0.10 |
ENSDART00000101601
|
cth1
|
cysteine three histidine 1 |
| chr23_+_27352072 | 0.10 |
ENSDART00000144762
|
si:dkey-9p24.5
|
si:dkey-9p24.5 |
| chr1_-_59169815 | 0.10 |
ENSDART00000100163
|
wu:fk65c09
|
wu:fk65c09 |
| chr25_+_15647750 | 0.10 |
ENSDART00000137375
|
spon1b
|
spondin 1b |
| chr2_+_33747509 | 0.10 |
ENSDART00000134187
|
si:dkey-31m5.5
|
si:dkey-31m5.5 |
| chr3_-_36419641 | 0.10 |
ENSDART00000173545
|
cog1
|
component of oligomeric golgi complex 1 |
| chr1_+_27153859 | 0.10 |
ENSDART00000180184
|
bnc2
|
basonuclin 2 |
| chr5_-_16475374 | 0.10 |
ENSDART00000134274
ENSDART00000136004 |
piwil2
|
piwi-like RNA-mediated gene silencing 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx3-1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 0.5 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.1 | 0.5 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.1 | 0.3 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.1 | 0.2 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.1 | 0.3 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.1 | 0.4 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.1 | 0.2 | GO:0006824 | cobalt ion transport(GO:0006824) cobalamin transport(GO:0015889) |
| 0.1 | 0.2 | GO:0019556 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.1 | 0.2 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 0.4 | GO:0055014 | ventricular cardiac myofibril assembly(GO:0055005) atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 0.4 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.1 | 0.2 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.1 | 0.3 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.0 | 0.2 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.0 | 0.2 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.0 | 0.3 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.2 | GO:0042762 | regulation of sulfur metabolic process(GO:0042762) |
| 0.0 | 0.2 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.0 | 0.3 | GO:1901798 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.0 | 0.3 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.2 | GO:0006772 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.0 | 0.2 | GO:0071938 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.0 | 0.5 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.4 | GO:0010717 | regulation of epithelial to mesenchymal transition(GO:0010717) |
| 0.0 | 0.9 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.1 | GO:1902292 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.0 | 0.1 | GO:0006601 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.0 | 0.1 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 | 0.5 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 0.4 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.1 | GO:1902024 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.0 | 0.1 | GO:2000638 | response to sterol(GO:0036314) cellular response to sterol(GO:0036315) SREBP-SCAP complex retention in endoplasmic reticulum(GO:0036316) regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0072003 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.0 | 0.5 | GO:0035141 | medial fin morphogenesis(GO:0035141) |
| 0.0 | 0.1 | GO:0031106 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.1 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.2 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.6 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.1 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.0 | 0.3 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.4 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.1 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.0 | 0.2 | GO:2000144 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 2.1 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.5 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.1 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.2 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.2 | GO:0006448 | regulation of translational elongation(GO:0006448) |
| 0.0 | 0.1 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.0 | 0.4 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.2 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.4 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.1 | GO:0044364 | neutrophil mediated immunity(GO:0002446) killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) disruption of cells of other organism involved in symbiotic interaction(GO:0051818) killing of cells in other organism involved in symbiotic interaction(GO:0051883) |
| 0.0 | 0.4 | GO:0007568 | aging(GO:0007568) |
| 0.0 | 0.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.2 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.2 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.0 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.1 | GO:0046314 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 0.1 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 0.0 | GO:0060295 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.1 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.2 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.1 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 0.2 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.0 | 0.5 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.2 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.4 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.2 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.4 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.2 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.2 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.2 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.3 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.1 | 0.5 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 1.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.4 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 0.2 | GO:0050333 | thiamin-triphosphatase activity(GO:0050333) |
| 0.1 | 0.4 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.1 | GO:0031530 | gonadotropin hormone-releasing hormone activity(GO:0005183) gonadotropin-releasing hormone receptor binding(GO:0031530) |
| 0.0 | 0.2 | GO:0005542 | folic acid binding(GO:0005542) ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.4 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.6 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.3 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.0 | 0.1 | GO:0034246 | core DNA-dependent RNA polymerase binding promoter specificity activity(GO:0000996) mitochondrial RNA polymerase binding promoter specificity activity(GO:0034246) |
| 0.0 | 0.5 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.2 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.2 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 0.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.2 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 2.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.0 | 0.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.2 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.2 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.0 | 0.1 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.5 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.1 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.5 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.2 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.4 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.2 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.4 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.2 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.0 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.2 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.2 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.2 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 0.6 | REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | Genes involved in p53-Independent G1/S DNA damage checkpoint |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.4 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |