Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
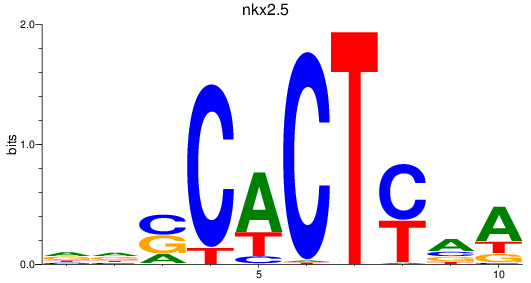
Results for nkx2.5
Z-value: 0.35
Transcription factors associated with nkx2.5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx2.5
|
ENSDARG00000018004 | NK2 homeobox 5 |
|
nkx2.5
|
ENSDARG00000116714 | NK2 homeobox 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nkx2.5 | dr11_v1_chr14_+_24241241_24241241 | -0.45 | 5.5e-02 | Click! |
Activity profile of nkx2.5 motif
Sorted Z-values of nkx2.5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_16764751 | 0.92 |
ENSDART00000113862
|
zgc:174154
|
zgc:174154 |
| chr12_-_16636627 | 0.90 |
ENSDART00000128811
|
si:dkey-239j18.3
|
si:dkey-239j18.3 |
| chr14_+_16036139 | 0.45 |
ENSDART00000190733
|
prelid1a
|
PRELI domain containing 1a |
| chr5_+_250078 | 0.39 |
ENSDART00000127504
|
trabd2a
|
TraB domain containing 2A |
| chr3_-_13599482 | 0.39 |
ENSDART00000166639
|
tufm
|
Tu translation elongation factor, mitochondrial |
| chr12_-_17479078 | 0.39 |
ENSDART00000079115
|
papss2b
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2b |
| chr23_+_25292147 | 0.39 |
ENSDART00000131486
|
pa2g4b
|
proliferation-associated 2G4, b |
| chr12_+_5102670 | 0.34 |
ENSDART00000166600
|
cep55l
|
centrosomal protein 55 like |
| chr20_-_25643667 | 0.33 |
ENSDART00000137457
|
si:dkeyp-117h8.4
|
si:dkeyp-117h8.4 |
| chr4_+_37406676 | 0.32 |
ENSDART00000130981
|
si:ch73-134f24.1
|
si:ch73-134f24.1 |
| chr20_-_26588736 | 0.32 |
ENSDART00000134337
|
exoc2
|
exocyst complex component 2 |
| chr8_-_28349859 | 0.31 |
ENSDART00000062671
|
tuba8l
|
tubulin, alpha 8 like |
| chr9_+_30294096 | 0.31 |
ENSDART00000026551
|
srpx
|
sushi-repeat containing protein, X-linked |
| chr6_+_58622831 | 0.30 |
ENSDART00000128793
|
sp7
|
Sp7 transcription factor |
| chr15_-_35246742 | 0.30 |
ENSDART00000131479
|
mff
|
mitochondrial fission factor |
| chr3_-_49514874 | 0.28 |
ENSDART00000167179
|
asf1ba
|
anti-silencing function 1Ba histone chaperone |
| chr25_-_11016675 | 0.27 |
ENSDART00000099572
|
mespab
|
mesoderm posterior ab |
| chr13_-_50614639 | 0.27 |
ENSDART00000170527
|
vent
|
ventral expressed homeobox |
| chr17_+_13088594 | 0.27 |
ENSDART00000193207
|
gemin2
|
gem (nuclear organelle) associated protein 2 |
| chr10_-_33265566 | 0.27 |
ENSDART00000063642
|
tbl2
|
transducin (beta)-like 2 |
| chr6_-_19341184 | 0.26 |
ENSDART00000168236
ENSDART00000167674 |
mif4gda
|
MIF4G domain containing a |
| chr15_-_47865063 | 0.26 |
ENSDART00000151600
|
h3f3b.1
|
H3 histone, family 3B.1 |
| chr15_+_823437 | 0.25 |
ENSDART00000155608
|
si:dkey-7i4.7
|
si:dkey-7i4.7 |
| chr6_+_58832155 | 0.25 |
ENSDART00000144842
|
dctn2
|
dynactin 2 (p50) |
| chr9_+_30475563 | 0.24 |
ENSDART00000133118
|
gja5a
|
gap junction protein, alpha 5a |
| chr10_-_29831944 | 0.24 |
ENSDART00000063923
ENSDART00000136264 |
zpr1
|
ZPR1 zinc finger |
| chr7_-_20464133 | 0.24 |
ENSDART00000078192
|
cnpy4
|
canopy4 |
| chr12_+_20587179 | 0.24 |
ENSDART00000170127
|
arsg
|
arylsulfatase G |
| chr15_-_17138640 | 0.24 |
ENSDART00000080777
|
mrpl28
|
mitochondrial ribosomal protein L28 |
| chr19_+_46240171 | 0.24 |
ENSDART00000162785
|
mapk15
|
mitogen-activated protein kinase 15 |
| chr7_-_67316903 | 0.24 |
ENSDART00000159408
|
psmd7
|
proteasome 26S subunit, non-ATPase 7 |
| chr12_-_13336703 | 0.24 |
ENSDART00000134356
|
lsm5
|
LSM5 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr2_+_30513887 | 0.24 |
ENSDART00000137048
|
march6
|
membrane-associated ring finger (C3HC4) 6 |
| chr8_-_22558773 | 0.23 |
ENSDART00000074309
|
porcnl
|
porcupine O-acyltransferase like |
| chr4_+_52356485 | 0.23 |
ENSDART00000170639
|
zgc:173705
|
zgc:173705 |
| chr7_+_40638210 | 0.23 |
ENSDART00000052236
|
mnx1
|
motor neuron and pancreas homeobox 1 |
| chr12_+_6098713 | 0.23 |
ENSDART00000139054
|
sgms1
|
sphingomyelin synthase 1 |
| chr13_-_6218248 | 0.23 |
ENSDART00000159052
|
si:zfos-1056e6.1
|
si:zfos-1056e6.1 |
| chr17_-_28797395 | 0.22 |
ENSDART00000134735
|
scfd1
|
sec1 family domain containing 1 |
| chr13_-_28610965 | 0.22 |
ENSDART00000043156
|
cyp17a1
|
cytochrome P450, family 17, subfamily A, polypeptide 1 |
| chr5_+_2002804 | 0.22 |
ENSDART00000064088
|
vkorc1l1
|
vitamin K epoxide reductase complex, subunit 1-like 1 |
| chr7_+_24390939 | 0.22 |
ENSDART00000087494
ENSDART00000125463 |
haus3
|
HAUS augmin-like complex, subunit 3 |
| chr16_-_31824525 | 0.21 |
ENSDART00000058737
|
cdc42l
|
cell division cycle 42, like |
| chr8_-_45759137 | 0.20 |
ENSDART00000123878
|
ppiaa
|
peptidylprolyl isomerase Aa (cyclophilin A) |
| chr11_+_39928828 | 0.20 |
ENSDART00000137516
ENSDART00000134082 |
vamp3
|
vesicle-associated membrane protein 3 (cellubrevin) |
| chr6_+_55428924 | 0.20 |
ENSDART00000018270
|
ncoa5
|
nuclear receptor coactivator 5 |
| chr20_+_34390196 | 0.20 |
ENSDART00000183596
|
trmt1l
|
tRNA methyltransferase 1-like |
| chr20_+_25904199 | 0.20 |
ENSDART00000016864
|
slc35f6
|
solute carrier family 35, member F6 |
| chr6_+_11990733 | 0.20 |
ENSDART00000151075
|
baz2ba
|
bromodomain adjacent to zinc finger domain, 2Ba |
| chr4_+_13901458 | 0.20 |
ENSDART00000137549
|
pphln1
|
periphilin 1 |
| chr5_-_26765188 | 0.20 |
ENSDART00000029450
|
rnf181
|
ring finger protein 181 |
| chr12_-_44072716 | 0.20 |
ENSDART00000162509
|
si:ch73-329n5.3
|
si:ch73-329n5.3 |
| chr22_+_25236888 | 0.19 |
ENSDART00000037286
|
zgc:172218
|
zgc:172218 |
| chr7_-_30639385 | 0.19 |
ENSDART00000173618
|
myo1ea
|
myosin IE, a |
| chr14_+_31493119 | 0.19 |
ENSDART00000006463
|
phf6
|
PHD finger protein 6 |
| chr4_-_77130289 | 0.19 |
ENSDART00000174380
|
CU467646.7
|
|
| chr3_+_29640837 | 0.19 |
ENSDART00000132298
|
eif3d
|
eukaryotic translation initiation factor 3, subunit D |
| chr3_+_35812040 | 0.19 |
ENSDART00000075903
ENSDART00000147712 |
crlf3
|
cytokine receptor-like factor 3 |
| chr12_+_22576404 | 0.19 |
ENSDART00000172053
|
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr6_+_58832323 | 0.19 |
ENSDART00000042595
|
dctn2
|
dynactin 2 (p50) |
| chr4_-_77125693 | 0.18 |
ENSDART00000174256
|
CU467646.3
|
|
| chr3_-_26805455 | 0.18 |
ENSDART00000180648
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr4_+_13428993 | 0.18 |
ENSDART00000067151
|
si:dkey-39a18.1
|
si:dkey-39a18.1 |
| chr8_-_20243389 | 0.18 |
ENSDART00000184904
|
acer1
|
alkaline ceramidase 1 |
| chr5_-_24543526 | 0.18 |
ENSDART00000046384
|
trmt2a
|
tRNA methyltransferase 2 homolog A |
| chr3_+_32571929 | 0.18 |
ENSDART00000151025
|
si:ch73-248e21.1
|
si:ch73-248e21.1 |
| chr15_+_24676905 | 0.18 |
ENSDART00000078014
ENSDART00000143137 |
poldip2
|
polymerase (DNA-directed), delta interacting protein 2 |
| chr7_+_16352924 | 0.18 |
ENSDART00000158972
|
mpped2a
|
metallophosphoesterase domain containing 2a |
| chr3_-_32079916 | 0.18 |
ENSDART00000040900
|
baxb
|
BCL2 associated X, apoptosis regulator b |
| chr20_-_53996193 | 0.18 |
ENSDART00000004756
|
hsp90aa1.1
|
heat shock protein 90, alpha (cytosolic), class A member 1, tandem duplicate 1 |
| chr20_+_14789305 | 0.18 |
ENSDART00000002463
|
tmed5
|
transmembrane p24 trafficking protein 5 |
| chr16_-_17200120 | 0.18 |
ENSDART00000147739
|
gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr12_+_17201522 | 0.18 |
ENSDART00000014536
ENSDART00000152452 |
rnls
|
renalase, FAD-dependent amine oxidase |
| chr22_-_19552796 | 0.18 |
ENSDART00000148088
ENSDART00000105485 |
si:dkey-78l4.14
|
si:dkey-78l4.14 |
| chr1_-_28860732 | 0.17 |
ENSDART00000177588
|
timmdc1
|
translocase of inner mitochondrial membrane domain containing 1 |
| chr2_-_37280028 | 0.17 |
ENSDART00000139459
|
nadkb
|
NAD kinase b |
| chr19_+_7292654 | 0.17 |
ENSDART00000140459
|
ccdc127b
|
coiled-coil domain containing 127b |
| chr4_-_77135076 | 0.17 |
ENSDART00000174184
|
zgc:173770
|
zgc:173770 |
| chr12_-_19862912 | 0.17 |
ENSDART00000145788
|
shisa9a
|
shisa family member 9a |
| chr6_-_25952848 | 0.16 |
ENSDART00000076997
ENSDART00000148748 |
lmo4b
|
LIM domain only 4b |
| chr24_-_23839647 | 0.16 |
ENSDART00000125190
|
rrs1
|
RRS1 ribosome biogenesis regulator homolog (S. cerevisiae) |
| chr5_+_47975758 | 0.16 |
ENSDART00000097429
|
BX470189.1
|
|
| chr22_+_2229964 | 0.16 |
ENSDART00000112582
|
znf1161
|
zinc finger protein 1161 |
| chr8_+_25893071 | 0.16 |
ENSDART00000078161
|
tmem115
|
transmembrane protein 115 |
| chr9_+_8396755 | 0.16 |
ENSDART00000043067
|
zgc:171776
|
zgc:171776 |
| chr22_-_5663354 | 0.16 |
ENSDART00000081774
|
ccdc51
|
coiled-coil domain containing 51 |
| chr22_-_4892937 | 0.16 |
ENSDART00000147002
|
si:ch73-256j6.4
|
si:ch73-256j6.4 |
| chr14_-_33297287 | 0.16 |
ENSDART00000045555
ENSDART00000138294 ENSDART00000075056 |
rab41
|
RAB41, member RAS oncogene family |
| chr18_-_29977431 | 0.16 |
ENSDART00000135357
|
si:ch211-220f16.2
|
si:ch211-220f16.2 |
| chr11_-_21031773 | 0.16 |
ENSDART00000065985
|
fmoda
|
fibromodulin a |
| chr19_-_29832876 | 0.16 |
ENSDART00000005119
|
eif3i
|
eukaryotic translation initiation factor 3, subunit I |
| chr13_-_31346392 | 0.15 |
ENSDART00000134343
|
rrp12
|
ribosomal RNA processing 12 homolog |
| chr6_-_42036685 | 0.15 |
ENSDART00000155174
|
GPR62 (1 of many)
|
si:dkeyp-111e5.4 |
| chr2_+_27010439 | 0.15 |
ENSDART00000030547
|
cdh7a
|
cadherin 7a |
| chr24_+_1294176 | 0.15 |
ENSDART00000106637
|
si:ch73-134f24.1
|
si:ch73-134f24.1 |
| chr17_+_46739693 | 0.15 |
ENSDART00000097810
|
pimr22
|
Pim proto-oncogene, serine/threonine kinase, related 22 |
| chr11_-_8208464 | 0.15 |
ENSDART00000161283
|
pimr203
|
Pim proto-oncogene, serine/threonine kinase, related 203 |
| chr1_-_55118745 | 0.14 |
ENSDART00000133915
|
sertad2a
|
SERTA domain containing 2a |
| chr25_+_36152215 | 0.14 |
ENSDART00000036147
|
irx5b
|
iroquois homeobox 5b |
| chr5_-_61638125 | 0.14 |
ENSDART00000134314
|
si:dkey-261j4.3
|
si:dkey-261j4.3 |
| chr5_+_18047111 | 0.14 |
ENSDART00000132164
|
hira
|
histone cell cycle regulator a |
| chr15_-_21155641 | 0.14 |
ENSDART00000061098
ENSDART00000046443 |
A2ML1 (1 of many)
|
si:dkey-105h12.2 |
| chr3_+_39600562 | 0.14 |
ENSDART00000134309
ENSDART00000007170 |
prpsap2
|
phosphoribosyl pyrophosphate synthetase-associated protein 2 |
| chr22_+_38762693 | 0.14 |
ENSDART00000015016
ENSDART00000150187 |
alpi.1
|
alkaline phosphatase, intestinal, tandem duplicate 1 |
| chr13_-_32899322 | 0.13 |
ENSDART00000133882
|
rock2a
|
rho-associated, coiled-coil containing protein kinase 2a |
| chr2_+_24352497 | 0.13 |
ENSDART00000134909
|
pimr68
|
Pim proto-oncogene, serine/threonine kinase, related 68 |
| chr3_-_111350 | 0.13 |
ENSDART00000171887
|
zgc:110249
|
zgc:110249 |
| chr4_+_33423119 | 0.13 |
ENSDART00000189413
|
znf1065
|
zinc finger protein 1065 |
| chr1_+_46598764 | 0.13 |
ENSDART00000053240
|
cab39l
|
calcium binding protein 39-like |
| chr22_+_2598998 | 0.13 |
ENSDART00000176665
|
CU570894.1
|
|
| chr7_-_67248829 | 0.13 |
ENSDART00000192442
|
znf143a
|
zinc finger protein 143a |
| chr25_-_29080063 | 0.13 |
ENSDART00000181911
ENSDART00000138087 |
cox5aa
|
cytochrome c oxidase subunit Vaa |
| chr16_-_9675982 | 0.13 |
ENSDART00000113724
|
mal2
|
mal, T cell differentiation protein 2 (gene/pseudogene) |
| chr3_-_15487111 | 0.13 |
ENSDART00000011320
|
nfatc2ip
|
nuclear factor of activated T cells 2 interacting protein |
| chr17_-_10043273 | 0.13 |
ENSDART00000156078
|
baz1a
|
bromodomain adjacent to zinc finger domain, 1A |
| chr2_-_8609653 | 0.13 |
ENSDART00000193354
ENSDART00000189489 ENSDART00000186144 |
si:ch211-71m22.1
|
si:ch211-71m22.1 |
| chr13_+_7242916 | 0.13 |
ENSDART00000184238
|
aifm2
|
apoptosis-inducing factor, mitochondrion-associated, 2 |
| chr8_-_7440364 | 0.12 |
ENSDART00000180659
|
hdac6
|
histone deacetylase 6 |
| chr16_+_33142734 | 0.12 |
ENSDART00000138244
|
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr5_+_24543862 | 0.12 |
ENSDART00000029699
|
atp6v0a2b
|
ATPase H+ transporting V0 subunit a2b |
| chr8_+_21114338 | 0.12 |
ENSDART00000002186
|
uck2a
|
uridine-cytidine kinase 2a |
| chr17_+_6538733 | 0.12 |
ENSDART00000193712
|
slc5a6b
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr4_+_45357558 | 0.12 |
ENSDART00000150769
|
si:ch211-162i8.5
|
si:ch211-162i8.5 |
| chr7_-_3669868 | 0.12 |
ENSDART00000172907
|
si:ch211-282j17.12
|
si:ch211-282j17.12 |
| chr22_+_25236657 | 0.12 |
ENSDART00000138012
|
zgc:172218
|
zgc:172218 |
| chr20_-_5369105 | 0.12 |
ENSDART00000114316
|
sptlc2b
|
serine palmitoyltransferase, long chain base subunit 2b |
| chr17_-_44249538 | 0.12 |
ENSDART00000008816
|
otx2b
|
orthodenticle homeobox 2b |
| chr5_-_39982694 | 0.12 |
ENSDART00000138458
|
pimr200
|
Pim proto-oncogene, serine/threonine kinase, related 200 |
| chr7_-_35083585 | 0.12 |
ENSDART00000192732
|
agrp
|
agouti related neuropeptide |
| chr14_-_16476863 | 0.11 |
ENSDART00000089021
|
canx
|
calnexin |
| chr22_+_5663529 | 0.11 |
ENSDART00000106141
|
tma7
|
translation machinery associated 7 homolog |
| chr24_+_15655233 | 0.11 |
ENSDART00000143160
|
fbxo15
|
F-box protein 15 |
| chr7_-_59311165 | 0.11 |
ENSDART00000171105
|
m1ap
|
meiosis 1 associated protein |
| chr14_+_49602 | 0.11 |
ENSDART00000035581
|
OTOP1
|
otopetrin 1 |
| chr13_-_8229977 | 0.11 |
ENSDART00000139728
|
si:ch211-250c4.4
|
si:ch211-250c4.4 |
| chr8_+_11471350 | 0.11 |
ENSDART00000092355
ENSDART00000136184 |
tjp2b
|
tight junction protein 2b (zona occludens 2) |
| chr17_+_30205258 | 0.11 |
ENSDART00000076596
ENSDART00000153795 |
spata17
|
spermatogenesis associated 17 |
| chr1_-_49950643 | 0.11 |
ENSDART00000138301
|
sgms2
|
sphingomyelin synthase 2 |
| chr22_+_1330477 | 0.11 |
ENSDART00000157567
|
CU207221.2
|
|
| chr11_-_36474306 | 0.11 |
ENSDART00000170678
ENSDART00000123591 |
usp48
|
ubiquitin specific peptidase 48 |
| chr22_+_25242322 | 0.11 |
ENSDART00000134628
|
si:ch211-226h8.8
|
si:ch211-226h8.8 |
| chr12_-_26423439 | 0.11 |
ENSDART00000113978
|
synpo2lb
|
synaptopodin 2-like b |
| chr22_-_18387059 | 0.11 |
ENSDART00000007769
|
tssk6
|
testis-specific serine kinase 6 |
| chr6_+_15373153 | 0.11 |
ENSDART00000155865
|
tmtops2a
|
teleost multiple tissue opsin 2a |
| chr6_+_55285578 | 0.11 |
ENSDART00000180183
|
zgc:109913
|
zgc:109913 |
| chr23_+_22291440 | 0.11 |
ENSDART00000185126
|
CR354612.1
|
|
| chr4_+_73606482 | 0.10 |
ENSDART00000150765
|
si:ch211-165i18.2
|
si:ch211-165i18.2 |
| chr11_-_34232906 | 0.10 |
ENSDART00000162150
|
lsg1
|
large 60S subunit nuclear export GTPase 1 |
| chr7_-_35083184 | 0.10 |
ENSDART00000100253
ENSDART00000135250 ENSDART00000173511 |
agrp
|
agouti related neuropeptide |
| chr5_-_13564961 | 0.10 |
ENSDART00000146827
|
si:ch211-230g14.3
|
si:ch211-230g14.3 |
| chr1_+_47178529 | 0.10 |
ENSDART00000158432
ENSDART00000074450 ENSDART00000137448 |
morc3b
|
MORC family CW-type zinc finger 3b |
| chr24_-_26945390 | 0.10 |
ENSDART00000123354
|
msl2b
|
male-specific lethal 2 homolog b (Drosophila) |
| chr10_+_8481660 | 0.10 |
ENSDART00000109559
|
BX682234.1
|
|
| chr11_-_12634017 | 0.10 |
ENSDART00000158286
ENSDART00000193090 |
CR450764.1
|
|
| chr16_+_44906324 | 0.10 |
ENSDART00000074960
|
cd22
|
cd22 molecule |
| chr12_+_19030391 | 0.10 |
ENSDART00000153927
|
si:ch73-139e5.2
|
si:ch73-139e5.2 |
| chr15_-_21132480 | 0.10 |
ENSDART00000078734
ENSDART00000157481 |
a2ml
|
alpha-2-macroglobulin-like |
| chr14_+_31493306 | 0.10 |
ENSDART00000138341
|
phf6
|
PHD finger protein 6 |
| chr8_+_48609521 | 0.10 |
ENSDART00000060765
|
nppb
|
natriuretic peptide B |
| chr16_-_41787421 | 0.09 |
ENSDART00000147210
|
si:dkey-199f5.8
|
si:dkey-199f5.8 |
| chr7_-_38664947 | 0.09 |
ENSDART00000100546
ENSDART00000112405 |
c6ast1
|
six-cysteine containing astacin protease 1 |
| chr21_+_19345558 | 0.09 |
ENSDART00000184733
|
hpse
|
heparanase |
| chr7_+_22585447 | 0.09 |
ENSDART00000149144
|
chrnb1l
|
cholinergic receptor, nicotinic, beta 1 (muscle) like |
| chr7_-_16562200 | 0.09 |
ENSDART00000169093
ENSDART00000173491 |
csrp3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chr20_-_46085840 | 0.09 |
ENSDART00000133714
|
taar12a
|
trace amine associated receptor 12a |
| chr6_-_40657653 | 0.09 |
ENSDART00000154359
|
ppil1
|
peptidylprolyl isomerase (cyclophilin)-like 1 |
| chr24_-_7587401 | 0.09 |
ENSDART00000093163
|
galnt11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr6_+_24817852 | 0.09 |
ENSDART00000165609
|
barhl2
|
BarH-like homeobox 2 |
| chr22_+_1481307 | 0.08 |
ENSDART00000177429
|
si:ch211-255f4.2
|
si:ch211-255f4.2 |
| chr18_+_33675664 | 0.08 |
ENSDART00000140043
|
si:dkey-47k20.8
|
si:dkey-47k20.8 |
| chr6_-_7726849 | 0.08 |
ENSDART00000151511
|
slc25a38b
|
solute carrier family 25, member 38b |
| chr16_+_9609721 | 0.08 |
ENSDART00000047920
|
enpp2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr5_-_42083363 | 0.08 |
ENSDART00000162596
|
cxcl11.5
|
chemokine (C-X-C motif) ligand 11, duplicate 5 |
| chr5_+_43782267 | 0.08 |
ENSDART00000130355
|
nos2a
|
nitric oxide synthase 2a, inducible |
| chr17_-_16342388 | 0.08 |
ENSDART00000017930
|
kcnk13a
|
potassium channel, subfamily K, member 13a |
| chr16_+_26612401 | 0.08 |
ENSDART00000145571
|
epb41l4b
|
erythrocyte membrane protein band 4.1 like 4B |
| chr23_-_24148646 | 0.08 |
ENSDART00000113598
|
arhgef19
|
Rho guanine nucleotide exchange factor (GEF) 19 |
| chr19_-_31372896 | 0.08 |
ENSDART00000046609
|
scin
|
scinderin |
| chr19_+_7292445 | 0.08 |
ENSDART00000026634
|
ccdc127b
|
coiled-coil domain containing 127b |
| chr21_+_27513859 | 0.08 |
ENSDART00000065420
|
pacs1a
|
phosphofurin acidic cluster sorting protein 1a |
| chr2_-_51303937 | 0.08 |
ENSDART00000164111
|
si:ch211-215e19.8
|
si:ch211-215e19.8 |
| chr2_+_42318012 | 0.07 |
ENSDART00000138137
|
ftr08
|
finTRIM family, member 8 |
| chr7_-_23777445 | 0.07 |
ENSDART00000173527
|
si:ch211-200p22.4
|
si:ch211-200p22.4 |
| chr24_-_2917540 | 0.07 |
ENSDART00000164776
|
fam69c
|
family with sequence similarity 69, member C |
| chr11_-_12379359 | 0.07 |
ENSDART00000187551
|
zgc:174353
|
zgc:174353 |
| chr4_+_33225161 | 0.07 |
ENSDART00000150851
|
si:dkey-247i3.5
|
si:dkey-247i3.5 |
| chr7_+_19817306 | 0.07 |
ENSDART00000044425
|
BX957278.1
|
|
| chr22_+_26793389 | 0.07 |
ENSDART00000165381
|
pimr69
|
Pim proto-oncogene, serine/threonine kinase, related 69 |
| chr2_+_33726862 | 0.07 |
ENSDART00000146745
|
si:dkey-31m5.5
|
si:dkey-31m5.5 |
| chr10_-_40410884 | 0.07 |
ENSDART00000150818
|
taar20n
|
trace amine associated receptor 20n |
| chr11_-_18017287 | 0.06 |
ENSDART00000155443
|
qrich1
|
glutamine-rich 1 |
| chr19_-_11966015 | 0.06 |
ENSDART00000123409
|
si:ch1073-296d18.1
|
si:ch1073-296d18.1 |
| chr9_-_34944604 | 0.06 |
ENSDART00000140563
ENSDART00000136812 |
dcun1d2a
|
DCN1, defective in cullin neddylation 1, domain containing 2a |
| chr11_-_12379541 | 0.06 |
ENSDART00000171717
|
zgc:174353
|
zgc:174353 |
| chr14_+_21828993 | 0.06 |
ENSDART00000144367
|
ctbp1
|
C-terminal binding protein 1 |
| chr1_+_58470287 | 0.06 |
ENSDART00000160245
ENSDART00000169945 |
si:ch73-236c18.7
|
si:ch73-236c18.7 |
| chr25_-_28674739 | 0.06 |
ENSDART00000067073
|
lrrc10
|
leucine rich repeat containing 10 |
| chr11_-_12363941 | 0.06 |
ENSDART00000192304
|
zgc:174353
|
zgc:174353 |
| chr15_+_11883804 | 0.06 |
ENSDART00000163255
|
gpr184
|
G protein-coupled receptor 184 |
| chr2_-_37280617 | 0.06 |
ENSDART00000190458
|
nadkb
|
NAD kinase b |
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx2.5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 0.4 | GO:0050427 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.1 | 0.2 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.1 | 0.2 | GO:0030237 | female sex determination(GO:0030237) male sex determination(GO:0030238) |
| 0.1 | 0.2 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.1 | 0.3 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.2 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.1 | 0.2 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.1 | 0.3 | GO:0033688 | regulation of osteoblast proliferation(GO:0033688) craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.1 | GO:0060031 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) mediolateral intercalation(GO:0060031) |
| 0.0 | 0.2 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 0.2 | GO:0045429 | regulation of nitric oxide biosynthetic process(GO:0045428) positive regulation of nitric oxide biosynthetic process(GO:0045429) positive regulation of reactive oxygen species biosynthetic process(GO:1903428) positive regulation of nitric oxide metabolic process(GO:1904407) |
| 0.0 | 0.2 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.0 | 0.4 | GO:0030719 | P granule organization(GO:0030719) |
| 0.0 | 0.1 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:1905072 | cardiac jelly development(GO:1905072) |
| 0.0 | 0.1 | GO:0051182 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.3 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.1 | GO:1904983 | transmembrane glycine transport from cytosol to mitochondrion(GO:1904983) |
| 0.0 | 0.2 | GO:0008343 | adult feeding behavior(GO:0008343) positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.2 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.1 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.2 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.3 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.0 | 0.1 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.0 | 0.1 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.0 | 0.2 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 0.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.2 | GO:0001836 | release of cytochrome c from mitochondria(GO:0001836) |
| 0.0 | 0.2 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.1 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.0 | 0.3 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.1 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.2 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.1 | GO:0031282 | regulation of cGMP metabolic process(GO:0030823) positive regulation of cGMP metabolic process(GO:0030825) regulation of cGMP biosynthetic process(GO:0030826) positive regulation of cGMP biosynthetic process(GO:0030828) regulation of guanylate cyclase activity(GO:0031282) positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.3 | GO:0014066 | regulation of phosphatidylinositol 3-kinase signaling(GO:0014066) |
| 0.0 | 0.0 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 0.0 | 0.1 | GO:0050848 | regulation of calcium-mediated signaling(GO:0050848) |
| 0.0 | 0.1 | GO:0008637 | apoptotic mitochondrial changes(GO:0008637) |
| 0.0 | 0.2 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.1 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.2 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.0 | 0.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.3 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.2 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.0 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.1 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.5 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.1 | 0.5 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 0.2 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.1 | 0.2 | GO:0031782 | type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.1 | 0.2 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.2 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.1 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.2 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0042903 | tubulin deacetylase activity(GO:0042903) acetylspermidine deacetylase activity(GO:0047611) |
| 0.0 | 0.1 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.6 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.2 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 1.9 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.2 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.0 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.0 | 0.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.2 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.1 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |