Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
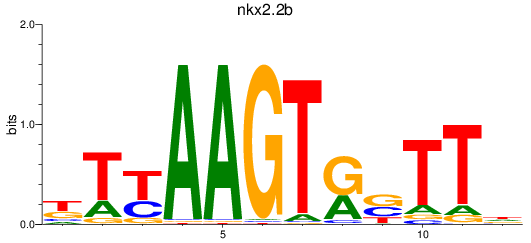
Results for nkx2.2b
Z-value: 0.44
Transcription factors associated with nkx2.2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx2.2b
|
ENSDARG00000101549 | NK2 homeobox 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nkx2.2b | dr11_v1_chr20_+_48782068_48782068 | 0.74 | 3.1e-04 | Click! |
Activity profile of nkx2.2b motif
Sorted Z-values of nkx2.2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_16595406 | 1.63 |
ENSDART00000166798
|
si:dkey-239j18.2
|
si:dkey-239j18.2 |
| chr12_-_16595177 | 1.57 |
ENSDART00000133962
|
si:dkey-239j18.2
|
si:dkey-239j18.2 |
| chr1_-_55248496 | 1.01 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr2_-_10877228 | 0.92 |
ENSDART00000138718
ENSDART00000034246 |
cdc7
|
cell division cycle 7 homolog (S. cerevisiae) |
| chr12_+_6002715 | 0.91 |
ENSDART00000114961
|
si:ch211-131k2.3
|
si:ch211-131k2.3 |
| chr9_-_33063083 | 0.80 |
ENSDART00000048550
|
si:ch211-125e6.5
|
si:ch211-125e6.5 |
| chr3_-_16000168 | 0.68 |
ENSDART00000160628
|
nme3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr1_+_5485799 | 0.67 |
ENSDART00000022307
|
atic
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase |
| chr3_-_15999501 | 0.67 |
ENSDART00000160668
|
nme3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr12_-_10476448 | 0.66 |
ENSDART00000106172
|
rac1a
|
Rac family small GTPase 1a |
| chr18_+_37272568 | 0.66 |
ENSDART00000132749
|
tmem123
|
transmembrane protein 123 |
| chr4_+_5333988 | 0.63 |
ENSDART00000129398
ENSDART00000163850 ENSDART00000067374 ENSDART00000150780 ENSDART00000150493 ENSDART00000150306 |
apex1
|
APEX nuclease (multifunctional DNA repair enzyme) 1 |
| chr23_-_19827411 | 0.63 |
ENSDART00000187964
|
haus7
|
HAUS augmin-like complex, subunit 7 |
| chr6_+_13201358 | 0.60 |
ENSDART00000190290
|
CT009620.1
|
|
| chr6_-_19341184 | 0.60 |
ENSDART00000168236
ENSDART00000167674 |
mif4gda
|
MIF4G domain containing a |
| chr6_+_22326624 | 0.59 |
ENSDART00000020333
|
rae1
|
ribonucleic acid export 1 |
| chr4_-_16451375 | 0.59 |
ENSDART00000192700
ENSDART00000128835 |
wu:fc23c09
|
wu:fc23c09 |
| chr7_+_21275152 | 0.58 |
ENSDART00000173612
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr12_-_4301234 | 0.58 |
ENSDART00000152377
ENSDART00000152521 |
ca15b
|
carbonic anhydrase XVb |
| chr20_+_2739804 | 0.57 |
ENSDART00000152655
|
syncrip
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr1_+_30723380 | 0.56 |
ENSDART00000127943
ENSDART00000062628 ENSDART00000127670 |
bora
|
bora, aurora kinase A activator |
| chr18_-_35407695 | 0.56 |
ENSDART00000191845
ENSDART00000141703 |
snrpa
|
small nuclear ribonucleoprotein polypeptide A |
| chr21_+_15603597 | 0.55 |
ENSDART00000138626
|
smarcb1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1b |
| chr25_+_28589669 | 0.55 |
ENSDART00000193901
|
tmem17
|
transmembrane protein 17 |
| chr23_+_38245610 | 0.55 |
ENSDART00000191386
|
znf217
|
zinc finger protein 217 |
| chr11_-_25257595 | 0.54 |
ENSDART00000123567
|
snai1a
|
snail family zinc finger 1a |
| chr22_+_2229964 | 0.54 |
ENSDART00000112582
|
znf1161
|
zinc finger protein 1161 |
| chr11_-_30636163 | 0.53 |
ENSDART00000140516
|
zgc:153665
|
zgc:153665 |
| chr7_-_22790630 | 0.53 |
ENSDART00000173496
|
si:ch211-15b10.6
|
si:ch211-15b10.6 |
| chr21_-_36619599 | 0.52 |
ENSDART00000065208
|
nop16
|
NOP16 nucleolar protein homolog (yeast) |
| chr4_+_5196469 | 0.52 |
ENSDART00000067386
|
rad51ap1
|
RAD51 associated protein 1 |
| chr13_-_30996072 | 0.51 |
ENSDART00000181661
|
wdfy4
|
WDFY family member 4 |
| chr15_+_37589698 | 0.50 |
ENSDART00000076066
ENSDART00000153894 ENSDART00000156298 |
lin37
|
lin-37 DREAM MuvB core complex component |
| chr16_+_20915319 | 0.49 |
ENSDART00000079383
|
hoxa9b
|
homeobox A9b |
| chr3_-_27647845 | 0.48 |
ENSDART00000151625
|
si:ch211-157c3.4
|
si:ch211-157c3.4 |
| chr7_-_26518086 | 0.48 |
ENSDART00000058913
|
eif4a1a
|
eukaryotic translation initiation factor 4A1A |
| chr20_+_2739531 | 0.48 |
ENSDART00000152269
|
syncrip
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr2_-_37956768 | 0.48 |
ENSDART00000034595
|
cbln10
|
cerebellin 10 |
| chr4_+_28356606 | 0.47 |
ENSDART00000192995
|
si:ch73-263o4.4
|
si:ch73-263o4.4 |
| chr14_+_10624911 | 0.47 |
ENSDART00000131295
|
nup62l
|
nucleoporin 62 like |
| chr4_+_33225161 | 0.46 |
ENSDART00000150851
|
si:dkey-247i3.5
|
si:dkey-247i3.5 |
| chr4_-_16876281 | 0.46 |
ENSDART00000016690
ENSDART00000044005 ENSDART00000042874 ENSDART00000125762 ENSDART00000185974 |
tmpoa
|
thymopoietin a |
| chr8_-_30779101 | 0.46 |
ENSDART00000062229
|
p2rx7
|
purinergic receptor P2X, ligand-gated ion channel, 7 |
| chr4_+_5334202 | 0.45 |
ENSDART00000150409
|
apex1
|
APEX nuclease (multifunctional DNA repair enzyme) 1 |
| chr18_-_27316599 | 0.45 |
ENSDART00000028294
|
zgc:56106
|
zgc:56106 |
| chr4_-_12477224 | 0.45 |
ENSDART00000027756
ENSDART00000182706 ENSDART00000127150 |
arhgef39
|
Rho guanine nucleotide exchange factor (GEF) 39 |
| chr18_+_26428829 | 0.45 |
ENSDART00000190779
ENSDART00000193226 ENSDART00000110746 |
blm
|
Bloom syndrome, RecQ helicase-like |
| chr6_+_13083146 | 0.45 |
ENSDART00000172158
|
l3hypdh
|
trans-L-3-hydroxyproline dehydratase |
| chr17_+_19481049 | 0.45 |
ENSDART00000024194
|
kif11
|
kinesin family member 11 |
| chr22_-_21676364 | 0.44 |
ENSDART00000183668
|
tle2b
|
transducin like enhancer of split 2b |
| chr4_-_20081621 | 0.44 |
ENSDART00000024647
|
dennd6b
|
DENN/MADD domain containing 6B |
| chr19_-_22328154 | 0.44 |
ENSDART00000090464
|
si:ch73-196l6.5
|
si:ch73-196l6.5 |
| chr5_-_26323137 | 0.43 |
ENSDART00000133823
|
arvcfb
|
ARVCF, delta catenin family member b |
| chr16_+_40340523 | 0.43 |
ENSDART00000102571
|
mettl6
|
methyltransferase like 6 |
| chr9_+_21722733 | 0.43 |
ENSDART00000102021
|
sox1a
|
SRY (sex determining region Y)-box 1a |
| chr15_-_20468302 | 0.42 |
ENSDART00000018514
|
dlc
|
deltaC |
| chr25_+_10485103 | 0.42 |
ENSDART00000104576
|
psmd13
|
proteasome 26S subunit, non-ATPase 13 |
| chr21_-_14826066 | 0.41 |
ENSDART00000067001
|
noc4l
|
nucleolar complex associated 4 homolog |
| chr16_-_14248892 | 0.41 |
ENSDART00000090329
|
rnf115
|
ring finger protein 115 |
| chr14_-_33297287 | 0.41 |
ENSDART00000045555
ENSDART00000138294 ENSDART00000075056 |
rab41
|
RAB41, member RAS oncogene family |
| chr14_+_14043793 | 0.41 |
ENSDART00000164376
|
rraga
|
Ras-related GTP binding A |
| chr20_+_51833030 | 0.41 |
ENSDART00000074330
|
taf1a
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, A |
| chr19_+_42231431 | 0.41 |
ENSDART00000102698
|
jtb
|
jumping translocation breakpoint |
| chr10_-_6494070 | 0.41 |
ENSDART00000171320
|
dcp2
|
decapping mRNA 2 |
| chr14_+_45608576 | 0.40 |
ENSDART00000173248
|
si:ch211-276i12.11
|
si:ch211-276i12.11 |
| chr4_+_6032640 | 0.40 |
ENSDART00000157487
|
tfec
|
transcription factor EC |
| chr12_+_36413886 | 0.40 |
ENSDART00000126325
|
si:ch211-250n8.1
|
si:ch211-250n8.1 |
| chr21_+_19547806 | 0.40 |
ENSDART00000159707
ENSDART00000184869 ENSDART00000181321 ENSDART00000058487 ENSDART00000058485 |
rai14
|
retinoic acid induced 14 |
| chr5_+_28497956 | 0.40 |
ENSDART00000191935
|
nfr
|
notochord formation related |
| chr7_-_71585065 | 0.40 |
ENSDART00000128678
|
mettl4
|
methyltransferase like 4 |
| chr14_-_26392475 | 0.39 |
ENSDART00000170614
|
b4galt7
|
xylosylprotein beta 1,4-galactosyltransferase, polypeptide 7 (galactosyltransferase I) |
| chr6_+_18544791 | 0.39 |
ENSDART00000167463
ENSDART00000169599 |
atad5b
|
ATPase family, AAA domain containing 5b |
| chr9_-_18911608 | 0.39 |
ENSDART00000138785
|
si:dkey-239h2.3
|
si:dkey-239h2.3 |
| chr10_+_44984946 | 0.39 |
ENSDART00000182520
|
h2afvb
|
H2A histone family, member Vb |
| chr17_-_10000339 | 0.39 |
ENSDART00000162893
|
snx6
|
sorting nexin 6 |
| chr6_+_49255706 | 0.39 |
ENSDART00000156866
|
si:dkey-183k8.2
|
si:dkey-183k8.2 |
| chr21_-_3007412 | 0.39 |
ENSDART00000190839
|
CKS2
|
zgc:86839 |
| chr2_+_38226145 | 0.39 |
ENSDART00000159396
|
si:ch211-14a17.11
|
si:ch211-14a17.11 |
| chr3_-_34136778 | 0.38 |
ENSDART00000131951
|
clpp
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr17_-_26610814 | 0.38 |
ENSDART00000133402
ENSDART00000016608 |
mrpl57
|
mitochondrial ribosomal protein L57 |
| chr14_-_25956804 | 0.38 |
ENSDART00000135627
ENSDART00000146022 ENSDART00000039660 |
sparc
|
secreted protein, acidic, cysteine-rich (osteonectin) |
| chr13_-_33317323 | 0.37 |
ENSDART00000110295
ENSDART00000144848 ENSDART00000136701 |
tmem234
|
transmembrane protein 234 |
| chr22_-_4892937 | 0.37 |
ENSDART00000147002
|
si:ch73-256j6.4
|
si:ch73-256j6.4 |
| chr17_+_6538733 | 0.37 |
ENSDART00000193712
|
slc5a6b
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr5_-_55914268 | 0.37 |
ENSDART00000014049
|
wdr36
|
WD repeat domain 36 |
| chr14_-_10617923 | 0.37 |
ENSDART00000133723
ENSDART00000131939 ENSDART00000136649 |
si:dkey-92i17.2
|
si:dkey-92i17.2 |
| chr7_-_8309505 | 0.36 |
ENSDART00000182530
|
f13a1b
|
coagulation factor XIII, A1 polypeptide b |
| chr13_+_33304187 | 0.36 |
ENSDART00000075826
ENSDART00000145295 |
dcdc2b
|
doublecortin domain containing 2B |
| chr7_-_67316903 | 0.36 |
ENSDART00000159408
|
psmd7
|
proteasome 26S subunit, non-ATPase 7 |
| chr18_-_22735002 | 0.36 |
ENSDART00000023721
|
nudt21
|
nudix hydrolase 21 |
| chr7_+_65296901 | 0.35 |
ENSDART00000162693
ENSDART00000161609 |
bola3
|
bolA family member 3 |
| chr10_-_44924289 | 0.35 |
ENSDART00000171267
|
tuba7l
|
tubulin, alpha 7 like |
| chr3_-_26805455 | 0.35 |
ENSDART00000180648
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr4_-_5333971 | 0.35 |
ENSDART00000067375
|
osgep
|
O-sialoglycoprotein endopeptidase |
| chr22_-_29922872 | 0.35 |
ENSDART00000020249
|
dusp5
|
dual specificity phosphatase 5 |
| chr2_-_10632431 | 0.35 |
ENSDART00000122709
|
mtf2
|
metal response element binding transcription factor 2 |
| chr1_+_8534698 | 0.34 |
ENSDART00000021504
|
smcr8b
|
Smith-Magenis syndrome chromosome region, candidate 8b |
| chr7_-_35083585 | 0.34 |
ENSDART00000192732
|
agrp
|
agouti related neuropeptide |
| chr24_+_41989108 | 0.34 |
ENSDART00000169725
|
zbtb14
|
zinc finger and BTB domain containing 14 |
| chr12_+_19138452 | 0.33 |
ENSDART00000141346
ENSDART00000066397 |
phf5a
|
PHD finger protein 5A |
| chr18_-_14691727 | 0.33 |
ENSDART00000010129
|
pdf
|
peptide deformylase, mitochondrial |
| chr11_+_42422371 | 0.33 |
ENSDART00000163780
|
dennd6aa
|
DENN/MADD domain containing 6Aa |
| chr7_+_37742299 | 0.33 |
ENSDART00000143300
|
brd7
|
bromodomain containing 7 |
| chr9_+_6255682 | 0.32 |
ENSDART00000149827
|
uxs1
|
UDP-glucuronate decarboxylase 1 |
| chr25_+_19008497 | 0.32 |
ENSDART00000104420
|
samm50
|
SAMM50 sorting and assembly machinery component |
| chr23_+_19813677 | 0.32 |
ENSDART00000139192
ENSDART00000142308 |
emd
|
emerin (Emery-Dreifuss muscular dystrophy) |
| chr4_+_38103421 | 0.31 |
ENSDART00000164164
|
znf1016
|
zinc finger protein 1016 |
| chr20_+_25712276 | 0.31 |
ENSDART00000121585
ENSDART00000185772 |
cep135
|
centrosomal protein 135 |
| chr4_+_69823638 | 0.31 |
ENSDART00000165786
|
znf1087
|
zinc finger protein 1087 |
| chr11_+_1565806 | 0.31 |
ENSDART00000137479
|
mybl2b
|
v-myb avian myeloblastosis viral oncogene homolog-like 2b |
| chr10_-_25343521 | 0.31 |
ENSDART00000166348
ENSDART00000009477 |
cct8
|
chaperonin containing TCP1, subunit 8 (theta) |
| chr3_+_12829566 | 0.31 |
ENSDART00000157672
|
si:ch211-8c17.2
|
si:ch211-8c17.2 |
| chr15_+_24746444 | 0.31 |
ENSDART00000032306
|
ywhae1
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide 1 |
| chr5_-_17601759 | 0.31 |
ENSDART00000138387
|
si:ch211-130h14.6
|
si:ch211-130h14.6 |
| chr6_-_42377307 | 0.31 |
ENSDART00000129302
|
emc3
|
ER membrane protein complex subunit 3 |
| chr25_-_16755340 | 0.31 |
ENSDART00000124729
ENSDART00000110859 |
ribc2
|
RIB43A domain with coiled-coils 2 |
| chr17_-_33412868 | 0.30 |
ENSDART00000187521
|
BX323819.1
|
|
| chr13_+_36595618 | 0.30 |
ENSDART00000022684
|
cnih1
|
cornichon family AMPA receptor auxiliary protein 1 |
| chr16_+_3185541 | 0.30 |
ENSDART00000024088
|
wdr21
|
WD repeat domain 21 |
| chr7_-_35083184 | 0.29 |
ENSDART00000100253
ENSDART00000135250 ENSDART00000173511 |
agrp
|
agouti related neuropeptide |
| chr1_-_46832880 | 0.29 |
ENSDART00000142406
|
si:ch73-160h15.3
|
si:ch73-160h15.3 |
| chr7_+_31132588 | 0.29 |
ENSDART00000173702
|
tjp1a
|
tight junction protein 1a |
| chr2_+_26498446 | 0.29 |
ENSDART00000078412
|
rps8a
|
ribosomal protein S8a |
| chr16_+_23913943 | 0.29 |
ENSDART00000175404
ENSDART00000129525 |
apoa4b.1
|
apolipoprotein A-IV b, tandem duplicate 1 |
| chr5_+_63390315 | 0.29 |
ENSDART00000124616
|
rab14
|
RAB14, member RAS oncogene family |
| chr24_+_5935377 | 0.29 |
ENSDART00000191989
ENSDART00000185932 ENSDART00000131768 |
abi1a
|
abl-interactor 1a |
| chr5_-_52752514 | 0.29 |
ENSDART00000190162
|
fam189a2
|
family with sequence similarity 189, member A2 |
| chr8_-_40205712 | 0.29 |
ENSDART00000158927
|
anapc5
|
anaphase promoting complex subunit 5 |
| chr15_-_16323750 | 0.29 |
ENSDART00000028500
|
nxn
|
nucleoredoxin |
| chr12_-_8511185 | 0.28 |
ENSDART00000179910
|
CR932364.1
|
|
| chr22_-_20720427 | 0.28 |
ENSDART00000105532
|
oaz1a
|
ornithine decarboxylase antizyme 1a |
| chr23_-_4225830 | 0.28 |
ENSDART00000170455
|
aar2
|
AAR2 splicing factor homolog (S. cerevisiae) |
| chr12_-_1031970 | 0.28 |
ENSDART00000105292
|
polr3e
|
polymerase (RNA) III (DNA directed) polypeptide E |
| chr15_+_31526225 | 0.28 |
ENSDART00000154456
|
wdr95
|
WD40 repeat domain 95 |
| chr17_-_20228610 | 0.28 |
ENSDART00000125758
|
ebf3b
|
early B cell factor 3b |
| chr20_+_492529 | 0.27 |
ENSDART00000141709
|
FO904980.1
|
|
| chr11_+_3308656 | 0.27 |
ENSDART00000082458
|
sarnp
|
SAP domain containing ribonucleoprotein |
| chr22_+_2100774 | 0.27 |
ENSDART00000106436
|
si:dkey-1b17.9
|
si:dkey-1b17.9 |
| chr3_+_60716904 | 0.27 |
ENSDART00000168280
|
foxj1a
|
forkhead box J1a |
| chr3_+_39579393 | 0.26 |
ENSDART00000055170
|
cln3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr22_+_22303053 | 0.26 |
ENSDART00000129975
|
scamp4
|
secretory carrier membrane protein 4 |
| chr3_-_26806032 | 0.26 |
ENSDART00000143710
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr2_-_53525896 | 0.26 |
ENSDART00000057053
|
im:7138239
|
im:7138239 |
| chr6_-_8465656 | 0.26 |
ENSDART00000178887
|
si:dkey-217d24.6
|
si:dkey-217d24.6 |
| chr18_-_40481028 | 0.26 |
ENSDART00000134177
|
zgc:101040
|
zgc:101040 |
| chr14_+_22132896 | 0.26 |
ENSDART00000138274
|
ccng1
|
cyclin G1 |
| chr3_-_15487111 | 0.26 |
ENSDART00000011320
|
nfatc2ip
|
nuclear factor of activated T cells 2 interacting protein |
| chr2_-_24402341 | 0.26 |
ENSDART00000155442
ENSDART00000088572 |
zgc:154006
|
zgc:154006 |
| chr4_-_32471434 | 0.26 |
ENSDART00000152013
ENSDART00000183621 |
znf1085
|
zinc finger protein 1085 |
| chr6_-_7439490 | 0.25 |
ENSDART00000188825
|
fkbp11
|
FK506 binding protein 11 |
| chr14_+_22132388 | 0.25 |
ENSDART00000109065
|
ccng1
|
cyclin G1 |
| chr14_-_45604667 | 0.25 |
ENSDART00000167016
|
si:ch211-276i12.9
|
si:ch211-276i12.9 |
| chr7_-_67317119 | 0.25 |
ENSDART00000183915
|
psmd7
|
proteasome 26S subunit, non-ATPase 7 |
| chr25_-_29134000 | 0.25 |
ENSDART00000172027
ENSDART00000190447 |
parp6b
|
poly (ADP-ribose) polymerase family, member 6b |
| chr8_-_18367736 | 0.25 |
ENSDART00000061679
|
dmap1
|
DNA methyltransferase 1 associated protein 1 |
| chr11_+_24900123 | 0.25 |
ENSDART00000044987
ENSDART00000148023 |
timm17a
|
translocase of inner mitochondrial membrane 17 homolog A (yeast) |
| chr2_-_22530969 | 0.25 |
ENSDART00000159641
|
znf644a
|
zinc finger protein 644a |
| chr16_+_5926520 | 0.25 |
ENSDART00000162229
|
ulk4
|
unc-51 like kinase 4 |
| chr14_-_21097574 | 0.24 |
ENSDART00000186803
|
rnf20
|
ring finger protein 20, E3 ubiquitin protein ligase |
| chr4_+_26449406 | 0.24 |
ENSDART00000183291
|
CU137645.3
|
|
| chr14_+_12169979 | 0.24 |
ENSDART00000129953
|
rhogd
|
ras homolog gene family, member Gd |
| chr3_-_7524363 | 0.24 |
ENSDART00000162970
|
znf1001
|
zinc finger protein 1001 |
| chr12_+_36109507 | 0.24 |
ENSDART00000175409
|
map2k6
|
mitogen-activated protein kinase kinase 6 |
| chr7_-_38087865 | 0.24 |
ENSDART00000052366
|
cebpa
|
CCAAT/enhancer binding protein (C/EBP), alpha |
| chr19_+_627899 | 0.24 |
ENSDART00000148508
|
tert
|
telomerase reverse transcriptase |
| chr24_-_30263301 | 0.24 |
ENSDART00000162328
|
snx7
|
sorting nexin 7 |
| chr2_-_3038904 | 0.24 |
ENSDART00000186795
|
guk1a
|
guanylate kinase 1a |
| chr13_-_22699024 | 0.24 |
ENSDART00000016946
|
glud1a
|
glutamate dehydrogenase 1a |
| chr6_+_27923054 | 0.24 |
ENSDART00000136833
ENSDART00000145533 |
cep63
|
centrosomal protein 63 |
| chr4_+_59234719 | 0.23 |
ENSDART00000170724
ENSDART00000192143 ENSDART00000109914 |
znf1086
|
zinc finger protein 1086 |
| chr10_-_27197044 | 0.23 |
ENSDART00000137928
|
auts2a
|
autism susceptibility candidate 2a |
| chr5_-_67499279 | 0.23 |
ENSDART00000128050
|
si:dkey-251i10.3
|
si:dkey-251i10.3 |
| chr2_-_21618629 | 0.23 |
ENSDART00000027587
|
ralab
|
v-ral simian leukemia viral oncogene homolog Ab (ras related) |
| chr23_+_1078072 | 0.23 |
ENSDART00000159263
ENSDART00000053527 |
slc34a2b
|
solute carrier family 34 (type II sodium/phosphate cotransporter), member 2b |
| chr21_+_25793970 | 0.23 |
ENSDART00000101217
|
CLDN4 (1 of many)
|
zgc:136892 |
| chr6_-_13408680 | 0.23 |
ENSDART00000151566
|
fmnl2b
|
formin-like 2b |
| chr4_-_41269844 | 0.23 |
ENSDART00000186177
|
CR388165.2
|
|
| chr14_-_42997145 | 0.23 |
ENSDART00000172801
|
pcdh10b
|
protocadherin 10b |
| chr4_+_62184754 | 0.23 |
ENSDART00000168844
|
si:dkeyp-35e5.9
|
si:dkeyp-35e5.9 |
| chr5_-_28052883 | 0.23 |
ENSDART00000188791
|
zgc:113436
|
zgc:113436 |
| chr15_-_684911 | 0.22 |
ENSDART00000161685
|
zgc:171901
|
zgc:171901 |
| chr12_+_29236274 | 0.22 |
ENSDART00000006505
|
mxtx2
|
mix-type homeobox gene 2 |
| chr11_+_31285127 | 0.22 |
ENSDART00000160154
|
si:dkey-238i5.2
|
si:dkey-238i5.2 |
| chr2_+_22531185 | 0.22 |
ENSDART00000171959
|
hfm1
|
HFM1, ATP-dependent DNA helicase homolog (S. cerevisiae) |
| chr2_+_2967255 | 0.22 |
ENSDART00000167649
ENSDART00000166449 |
pik3r3a
|
phosphoinositide-3-kinase, regulatory subunit 3a (gamma) |
| chr6_+_55212928 | 0.22 |
ENSDART00000083668
|
manbal
|
mannosidase beta like |
| chr5_+_3927989 | 0.22 |
ENSDART00000030125
|
znhit3
|
zinc finger, HIT-type containing 3 |
| chr7_-_44704910 | 0.22 |
ENSDART00000037850
|
dync1li2
|
dynein, cytoplasmic 1, light intermediate chain 2 |
| chr3_+_18807524 | 0.22 |
ENSDART00000055757
|
tnpo2
|
transportin 2 (importin 3, karyopherin beta 2b) |
| chr16_-_45042236 | 0.22 |
ENSDART00000058383
|
tmem147
|
transmembrane protein 147 |
| chr11_-_7411436 | 0.21 |
ENSDART00000167312
|
adgrl4
|
adhesion G protein-coupled receptor L4 |
| chr1_+_33328857 | 0.21 |
ENSDART00000137151
|
mxra5a
|
matrix-remodelling associated 5a |
| chr18_-_43866001 | 0.21 |
ENSDART00000150218
|
treh
|
trehalase (brush-border membrane glycoprotein) |
| chr10_-_41302841 | 0.21 |
ENSDART00000020297
ENSDART00000160174 ENSDART00000183850 ENSDART00000169493 |
brf2
|
BRF2, RNA polymerase III transcription initiation factor |
| chr15_-_37589600 | 0.21 |
ENSDART00000154641
|
proser3
|
proline and serine rich 3 |
| chr13_+_8840772 | 0.21 |
ENSDART00000059321
|
epcam
|
epithelial cell adhesion molecule |
| chr12_+_30726425 | 0.21 |
ENSDART00000153275
|
slc35f3a
|
solute carrier family 35, member F3a |
| chr9_+_55056310 | 0.21 |
ENSDART00000047080
|
gpr143
|
G protein-coupled receptor 143 |
| chr6_-_13200585 | 0.21 |
ENSDART00000185321
|
eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr2_-_1514001 | 0.21 |
ENSDART00000057736
|
c8b
|
complement component 8, beta polypeptide |
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx2.2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.1 | 0.4 | GO:2000425 | regulation of apoptotic cell clearance(GO:2000425) |
| 0.1 | 1.0 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 0.4 | GO:1990120 | messenger ribonucleoprotein complex assembly(GO:1990120) |
| 0.1 | 1.4 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.1 | 0.5 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.1 | 0.3 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.1 | 0.4 | GO:0015887 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.1 | 0.3 | GO:0060296 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.1 | 1.3 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.1 | 0.5 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 0.2 | GO:0007571 | age-dependent general metabolic decline(GO:0007571) |
| 0.1 | 0.6 | GO:0008343 | adult feeding behavior(GO:0008343) positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 0.3 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.1 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 0.4 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.1 | 0.3 | GO:0090387 | phagosome maturation involved in apoptotic cell clearance(GO:0090386) phagolysosome assembly involved in apoptotic cell clearance(GO:0090387) |
| 0.1 | 0.2 | GO:0005991 | trehalose metabolic process(GO:0005991) |
| 0.1 | 0.2 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.1 | 0.3 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 0.2 | GO:0021693 | cerebellar Purkinje cell layer structural organization(GO:0021693) cerebellar cortex structural organization(GO:0021698) |
| 0.1 | 0.3 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.1 | 0.7 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 0.2 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 0.9 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 0.4 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.1 | 0.4 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 0.2 | GO:0003156 | regulation of organ formation(GO:0003156) |
| 0.1 | 0.9 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.1 | 0.3 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 1.1 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.1 | 0.5 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.5 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.2 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.3 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.0 | 0.1 | GO:1902410 | mitotic cytokinetic process(GO:1902410) mitotic cleavage furrow formation(GO:1903673) |
| 0.0 | 0.3 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.1 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.0 | 0.1 | GO:0060785 | regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.0 | 0.5 | GO:0033198 | response to ATP(GO:0033198) |
| 0.0 | 0.3 | GO:0015809 | arginine transport(GO:0015809) |
| 0.0 | 0.1 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.0 | 0.4 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.2 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.2 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.0 | 0.3 | GO:0006478 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.2 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.2 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.2 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.9 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.1 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.2 | GO:0050848 | regulation of calcium-mediated signaling(GO:0050848) |
| 0.0 | 0.2 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.6 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.3 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.6 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.0 | 0.7 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.3 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.5 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.0 | 0.2 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.4 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.8 | GO:0002183 | cytoplasmic translational initiation(GO:0002183) |
| 0.0 | 0.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.6 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.3 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.2 | GO:0009146 | dGTP catabolic process(GO:0006203) purine nucleoside triphosphate catabolic process(GO:0009146) purine deoxyribonucleoside triphosphate catabolic process(GO:0009217) dGTP metabolic process(GO:0046070) |
| 0.0 | 0.1 | GO:0031284 | regulation of cGMP metabolic process(GO:0030823) positive regulation of cGMP metabolic process(GO:0030825) regulation of cGMP biosynthetic process(GO:0030826) positive regulation of cGMP biosynthetic process(GO:0030828) regulation of guanylate cyclase activity(GO:0031282) positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.6 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0032096 | negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.1 | GO:0045901 | positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.3 | GO:0006596 | polyamine biosynthetic process(GO:0006596) |
| 0.0 | 0.3 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 0.2 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.3 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.0 | 0.4 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 0.5 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.1 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.0 | 0.2 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.4 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.3 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.2 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 1.1 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.2 | GO:0009303 | rRNA transcription(GO:0009303) swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.0 | 0.1 | GO:0006691 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.0 | 0.2 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.2 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 0.0 | 0.5 | GO:0006405 | RNA export from nucleus(GO:0006405) |
| 0.0 | 0.2 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.8 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.2 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.1 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.0 | 0.2 | GO:0010390 | histone monoubiquitination(GO:0010390) |
| 0.0 | 0.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.4 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.1 | GO:0006537 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.0 | 0.4 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.4 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0030689 | Noc complex(GO:0030689) |
| 0.1 | 0.4 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 0.4 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 1.0 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 0.6 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 0.3 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 0.4 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 0.2 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.1 | 0.6 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.6 | GO:0035060 | brahma complex(GO:0035060) |
| 0.1 | 0.5 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 0.4 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.3 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.6 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.3 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 0.4 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:0070743 | interleukin-12 complex(GO:0043514) interleukin-23 complex(GO:0070743) |
| 0.0 | 0.6 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 0.5 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 1.0 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.3 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.6 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.2 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.0 | 0.6 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.2 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.3 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.2 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.6 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.2 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 4.0 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 0.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.3 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.5 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.7 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.5 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0008311 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.2 | 0.6 | GO:0031782 | type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.1 | 0.5 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 0.3 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.1 | 0.7 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.1 | 1.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.4 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.1 | 0.6 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.2 | GO:0003721 | telomerase RNA reverse transcriptase activity(GO:0003721) |
| 0.1 | 0.4 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.1 | 0.3 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 0.2 | GO:0004555 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 0.1 | 0.2 | GO:0072545 | tyrosine binding(GO:0072545) |
| 0.1 | 0.6 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.1 | 0.3 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.3 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.1 | 0.6 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.9 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 1.5 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 0.4 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.1 | 0.5 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.3 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.4 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.2 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.5 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.1 | GO:0019972 | interleukin-12 binding(GO:0019972) interleukin-12 alpha subunit binding(GO:0042164) |
| 0.0 | 0.4 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.3 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 0.3 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.1 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.6 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.1 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.1 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.4 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.0 | 0.5 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 0.4 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 0.2 | GO:0016793 | dGTPase activity(GO:0008832) triphosphoric monoester hydrolase activity(GO:0016793) |
| 0.0 | 0.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.3 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.3 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 3.4 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.3 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 1.2 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.2 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.1 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.0 | 0.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.6 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.3 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.6 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.2 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.1 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.1 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.9 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.2 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.5 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.5 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.4 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 0.5 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.7 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.4 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 1.1 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 0.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 1.0 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.2 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.4 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.4 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.4 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.6 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.2 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.3 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.2 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.1 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.3 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.6 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.4 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.2 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.5 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.9 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.2 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |