Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
Results for nkx2.2a
Z-value: 0.74
Transcription factors associated with nkx2.2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx2.2a
|
ENSDARG00000053298 | NK2 homeobox 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nkx2.2a | dr11_v1_chr17_-_42213822_42213822 | -0.68 | 1.3e-03 | Click! |
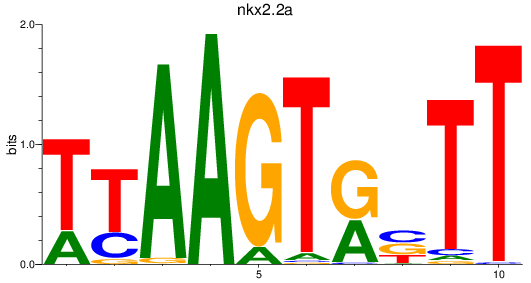
Activity profile of nkx2.2a motif
Sorted Z-values of nkx2.2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_32838110 | 2.23 |
ENSDART00000158077
|
arr3b
|
arrestin 3b, retinal (X-arrestin) |
| chr25_-_7974494 | 1.65 |
ENSDART00000171446
|
hal
|
histidine ammonia-lyase |
| chr7_+_50849142 | 1.62 |
ENSDART00000073806
|
pcolceb
|
procollagen C-endopeptidase enhancer b |
| chr22_+_20720808 | 1.31 |
ENSDART00000171321
|
si:dkey-211f22.5
|
si:dkey-211f22.5 |
| chr19_+_40856807 | 1.30 |
ENSDART00000139083
|
gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr24_-_7699356 | 1.24 |
ENSDART00000013117
|
syt5b
|
synaptotagmin Vb |
| chr20_-_34090740 | 1.06 |
ENSDART00000062539
ENSDART00000008140 |
pdcb
|
phosducin b |
| chr14_+_32837914 | 1.05 |
ENSDART00000158888
|
arr3b
|
arrestin 3b, retinal (X-arrestin) |
| chr16_-_11798994 | 1.02 |
ENSDART00000135408
|
cnfn
|
cornifelin |
| chr9_-_22158784 | 1.02 |
ENSDART00000167850
|
crygm2d14
|
crystallin, gamma M2d14 |
| chr19_-_12315693 | 1.00 |
ENSDART00000151158
|
ncaldb
|
neurocalcin delta b |
| chr9_-_22158325 | 1.00 |
ENSDART00000114568
|
crygm2d16
|
crystallin, gamma M2d16 |
| chr4_-_72609735 | 0.97 |
ENSDART00000174299
ENSDART00000159227 |
si:cabz01054394.6
|
si:cabz01054394.6 |
| chr12_-_19865585 | 0.93 |
ENSDART00000066386
|
shisa9a
|
shisa family member 9a |
| chr1_+_15137901 | 0.93 |
ENSDART00000111475
|
pcdh7a
|
protocadherin 7a |
| chr19_-_12322356 | 0.92 |
ENSDART00000016128
|
ncaldb
|
neurocalcin delta b |
| chr5_+_54685175 | 0.92 |
ENSDART00000115016
|
pmchl
|
pro-melanin-concentrating hormone, like |
| chr6_+_49255706 | 0.91 |
ENSDART00000156866
|
si:dkey-183k8.2
|
si:dkey-183k8.2 |
| chr7_-_38698583 | 0.91 |
ENSDART00000173900
ENSDART00000126737 |
cd59
|
CD59 molecule (CD59 blood group) |
| chr2_+_5948534 | 0.89 |
ENSDART00000124324
ENSDART00000176461 |
slc1a7a
|
solute carrier family 1 (glutamate transporter), member 7a |
| chr13_+_47710434 | 0.89 |
ENSDART00000188724
|
TMEM87B
|
transmembrane protein 87B |
| chr9_-_9998087 | 0.88 |
ENSDART00000124423
|
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr5_+_72087619 | 0.86 |
ENSDART00000062885
|
oxt
|
oxytocin |
| chr19_+_9344171 | 0.85 |
ENSDART00000133447
ENSDART00000104622 |
si:ch211-288g17.4
|
si:ch211-288g17.4 |
| chr12_-_3237561 | 0.83 |
ENSDART00000164665
|
si:ch1073-13h15.3
|
si:ch1073-13h15.3 |
| chr11_-_11518469 | 0.77 |
ENSDART00000104254
|
krt15
|
keratin 15 |
| chr5_-_23277939 | 0.75 |
ENSDART00000003514
|
plp1b
|
proteolipid protein 1b |
| chr25_+_20077225 | 0.74 |
ENSDART00000136543
|
tnni4b.1
|
troponin I4b, tandem duplicate 1 |
| chr6_+_4872883 | 0.74 |
ENSDART00000186730
ENSDART00000092290 ENSDART00000151674 |
pcdh9
|
protocadherin 9 |
| chr5_-_47727819 | 0.73 |
ENSDART00000165249
|
cox7c
|
cytochrome c oxidase, subunit VIIc |
| chr20_-_30920356 | 0.71 |
ENSDART00000022951
|
kif25
|
kinesin family member 25 |
| chr12_+_16440708 | 0.69 |
ENSDART00000113810
|
ankrd1b
|
ankyrin repeat domain 1b (cardiac muscle) |
| chr2_-_15318786 | 0.69 |
ENSDART00000135851
|
tecrl2b
|
trans-2,3-enoyl-CoA reductase-like 2b |
| chr1_-_22735924 | 0.68 |
ENSDART00000134831
|
prom1b
|
prominin 1 b |
| chr23_-_39784368 | 0.68 |
ENSDART00000110282
|
si:ch211-286f9.2
|
si:ch211-286f9.2 |
| chr11_-_11471857 | 0.66 |
ENSDART00000030103
|
krt94
|
keratin 94 |
| chr7_-_13884610 | 0.65 |
ENSDART00000006897
|
rlbp1a
|
retinaldehyde binding protein 1a |
| chr21_-_42055872 | 0.65 |
ENSDART00000144767
|
gabra1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr8_+_28065803 | 0.65 |
ENSDART00000178481
|
kcnd3
|
potassium voltage-gated channel, Shal-related subfamily, member 3 |
| chr13_-_23270576 | 0.63 |
ENSDART00000132828
|
si:dkey-103j14.5
|
si:dkey-103j14.5 |
| chr2_-_24269911 | 0.63 |
ENSDART00000099532
|
myh7
|
myosin heavy chain 7 |
| chr22_-_8006342 | 0.62 |
ENSDART00000162028
|
sc:d217
|
sc:d217 |
| chr19_+_37848830 | 0.61 |
ENSDART00000042276
ENSDART00000180872 |
nxph1
|
neurexophilin 1 |
| chr1_-_9231952 | 0.60 |
ENSDART00000166515
|
si:dkeyp-57d7.4
|
si:dkeyp-57d7.4 |
| chr16_+_30301539 | 0.59 |
ENSDART00000186018
|
LO017848.1
|
|
| chr20_-_34750045 | 0.58 |
ENSDART00000186130
|
znf395b
|
zinc finger protein 395b |
| chr6_+_36795225 | 0.57 |
ENSDART00000171504
|
si:ch73-29l19.1
|
si:ch73-29l19.1 |
| chr1_+_55662491 | 0.57 |
ENSDART00000152386
|
adgre8
|
adhesion G protein-coupled receptor E8 |
| chr5_-_30380593 | 0.57 |
ENSDART00000148039
|
snx19a
|
sorting nexin 19a |
| chr8_-_13823091 | 0.57 |
ENSDART00000177174
ENSDART00000137021 |
cabp4
|
calcium binding protein 4 |
| chr12_-_25150239 | 0.55 |
ENSDART00000038415
ENSDART00000135368 |
rhoq
|
ras homolog family member Q |
| chr13_+_28675686 | 0.55 |
ENSDART00000027213
|
inaa
|
internexin neuronal intermediate filament protein, alpha a |
| chr23_+_6795709 | 0.55 |
ENSDART00000149136
|
si:ch211-117c9.5
|
si:ch211-117c9.5 |
| chr6_+_21395051 | 0.54 |
ENSDART00000017774
|
cacng5a
|
calcium channel, voltage-dependent, gamma subunit 5a |
| chr19_-_10656667 | 0.54 |
ENSDART00000081379
ENSDART00000151456 ENSDART00000143271 ENSDART00000182126 |
olah
|
oleoyl-ACP hydrolase |
| chr25_-_32311048 | 0.53 |
ENSDART00000181806
ENSDART00000086334 |
CU372926.1
|
|
| chr9_-_11560427 | 0.53 |
ENSDART00000127942
ENSDART00000061442 |
cryba2b
|
crystallin, beta A2b |
| chr9_+_2762270 | 0.53 |
ENSDART00000123342
ENSDART00000001795 ENSDART00000177563 |
sp3a
|
sp3a transcription factor |
| chr8_-_1698155 | 0.53 |
ENSDART00000186159
|
CABZ01065417.1
|
|
| chr8_-_39739627 | 0.52 |
ENSDART00000135422
ENSDART00000067844 |
si:ch211-170d8.5
|
si:ch211-170d8.5 |
| chr13_-_29424454 | 0.51 |
ENSDART00000026765
|
slc18a3a
|
solute carrier family 18 (vesicular acetylcholine transporter), member 3a |
| chr7_+_41295974 | 0.50 |
ENSDART00000173568
ENSDART00000173544 |
si:dkey-86l18.10
|
si:dkey-86l18.10 |
| chr6_-_54121035 | 0.50 |
ENSDART00000183970
|
tusc2a
|
tumor suppressor candidate 2a |
| chr15_-_30714130 | 0.50 |
ENSDART00000156914
ENSDART00000154714 |
msi2b
|
musashi RNA-binding protein 2b |
| chr23_+_6795531 | 0.50 |
ENSDART00000092131
|
si:ch211-117c9.5
|
si:ch211-117c9.5 |
| chr5_+_9408901 | 0.49 |
ENSDART00000193364
|
FP236810.1
|
|
| chr1_+_41690402 | 0.47 |
ENSDART00000177298
|
fbxo41
|
F-box protein 41 |
| chr2_+_38924975 | 0.47 |
ENSDART00000109219
|
rem2
|
RAS (RAD and GEM)-like GTP binding 2 |
| chr4_-_17838179 | 0.46 |
ENSDART00000146931
|
avpr2l
|
arginine vasopressin receptor 2, like |
| chr13_-_31829786 | 0.46 |
ENSDART00000138667
|
sertad4
|
SERTA domain containing 4 |
| chr25_+_35375848 | 0.46 |
ENSDART00000155721
|
ano3
|
anoctamin 3 |
| chr6_+_11438972 | 0.46 |
ENSDART00000029314
|
col5a2b
|
collagen, type V, alpha 2b |
| chr20_-_34750363 | 0.45 |
ENSDART00000152845
|
znf395b
|
zinc finger protein 395b |
| chr8_+_8845932 | 0.45 |
ENSDART00000112028
|
si:ch211-180f4.1
|
si:ch211-180f4.1 |
| chr4_-_4387012 | 0.45 |
ENSDART00000191836
|
CU468826.3
|
Danio rerio U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit-related protein 1-like (LOC100331497), mRNA. |
| chr24_-_14212521 | 0.44 |
ENSDART00000130825
|
xkr9
|
XK, Kell blood group complex subunit-related family, member 9 |
| chr4_-_25836684 | 0.44 |
ENSDART00000142491
|
ndufa12
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12 |
| chr17_-_45040813 | 0.44 |
ENSDART00000075514
|
entpd5a
|
ectonucleoside triphosphate diphosphohydrolase 5a |
| chr1_-_1894722 | 0.44 |
ENSDART00000165669
|
si:ch211-132g1.3
|
si:ch211-132g1.3 |
| chr12_+_2631711 | 0.44 |
ENSDART00000114209
|
gdf10b
|
growth differentiation factor 10b |
| chr2_-_24270062 | 0.42 |
ENSDART00000192445
|
myh7
|
myosin heavy chain 7 |
| chr7_-_29571615 | 0.42 |
ENSDART00000019140
|
rorab
|
RAR-related orphan receptor A, paralog b |
| chr2_+_15203322 | 0.42 |
ENSDART00000144171
|
abca4b
|
ATP-binding cassette, sub-family A (ABC1), member 4b |
| chr3_+_56876280 | 0.42 |
ENSDART00000154197
|
amn
|
amnion associated transmembrane protein |
| chr2_+_51783120 | 0.41 |
ENSDART00000177559
|
crygn1
|
crystallin, gamma N1 |
| chr20_-_6812688 | 0.41 |
ENSDART00000170934
|
igfbp1a
|
insulin-like growth factor binding protein 1a |
| chr20_-_28349144 | 0.41 |
ENSDART00000179690
ENSDART00000188059 |
ino80
|
INO80 complex subunit |
| chr5_-_37117778 | 0.41 |
ENSDART00000149138
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr15_-_1198886 | 0.40 |
ENSDART00000063285
|
lxn
|
latexin |
| chr4_+_842010 | 0.40 |
ENSDART00000067461
|
si:ch211-152c2.3
|
si:ch211-152c2.3 |
| chr9_+_14010823 | 0.40 |
ENSDART00000143837
|
si:ch211-67e16.3
|
si:ch211-67e16.3 |
| chr9_-_51323545 | 0.40 |
ENSDART00000139316
|
slc4a10b
|
solute carrier family 4, sodium bicarbonate transporter, member 10b |
| chr7_-_16562200 | 0.40 |
ENSDART00000169093
ENSDART00000173491 |
csrp3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chr20_-_35512932 | 0.40 |
ENSDART00000137690
|
adgrf3b
|
adhesion G protein-coupled receptor F3b |
| chr7_-_28413224 | 0.39 |
ENSDART00000076502
|
rerglb
|
RERG/RAS-like b |
| chr22_-_11626014 | 0.38 |
ENSDART00000063133
ENSDART00000160085 |
gcga
|
glucagon a |
| chr9_+_42095220 | 0.38 |
ENSDART00000148317
ENSDART00000134431 |
pcbp3
|
poly(rC) binding protein 3 |
| chr11_-_3629201 | 0.38 |
ENSDART00000136577
ENSDART00000132121 |
itih3a
|
inter-alpha-trypsin inhibitor heavy chain 3a |
| chr17_+_30205258 | 0.38 |
ENSDART00000076596
ENSDART00000153795 |
spata17
|
spermatogenesis associated 17 |
| chr2_-_11662851 | 0.38 |
ENSDART00000145108
|
zgc:110130
|
zgc:110130 |
| chr2_+_31454562 | 0.37 |
ENSDART00000136360
|
TMEM236
|
si:dkey-32m20.1 |
| chr23_-_24856025 | 0.37 |
ENSDART00000142171
|
syt6a
|
synaptotagmin VIa |
| chr2_+_7192966 | 0.37 |
ENSDART00000142735
|
si:ch211-13f8.1
|
si:ch211-13f8.1 |
| chr25_+_8407892 | 0.37 |
ENSDART00000153536
|
muc5.2
|
mucin 5.2 |
| chr13_+_8840772 | 0.37 |
ENSDART00000059321
|
epcam
|
epithelial cell adhesion molecule |
| chr13_+_29352842 | 0.36 |
ENSDART00000113009
|
ogdhl
|
oxoglutarate dehydrogenase like |
| chr16_+_11652591 | 0.36 |
ENSDART00000133876
|
si:dkey-250k15.9
|
si:dkey-250k15.9 |
| chr14_+_36886950 | 0.36 |
ENSDART00000183719
|
si:ch211-132p1.3
|
si:ch211-132p1.3 |
| chr17_-_14726824 | 0.36 |
ENSDART00000162947
|
si:ch73-305o9.3
|
si:ch73-305o9.3 |
| chr3_-_8873716 | 0.36 |
ENSDART00000171798
|
znf1150
|
zinc finger protein 1150 |
| chr19_-_677713 | 0.36 |
ENSDART00000025146
|
slc6a19a.1
|
solute carrier family 6 (neutral amino acid transporter), member 19a, tandem duplicate 1 |
| chr17_-_28770800 | 0.35 |
ENSDART00000156485
|
opn6b
|
opsin 6, group member b |
| chr20_+_27020201 | 0.35 |
ENSDART00000126919
ENSDART00000016014 |
chga
|
chromogranin A |
| chr24_+_3963684 | 0.34 |
ENSDART00000182959
ENSDART00000185926 ENSDART00000167043 ENSDART00000033394 |
pfkpa
|
phosphofructokinase, platelet a |
| chr15_+_7054754 | 0.34 |
ENSDART00000149800
|
foxl2a
|
forkhead box L2a |
| chr3_+_52737565 | 0.34 |
ENSDART00000108639
|
gmip
|
GEM interacting protein |
| chr17_-_20143946 | 0.33 |
ENSDART00000138911
|
actn2b
|
actinin, alpha 2b |
| chr7_+_34231782 | 0.33 |
ENSDART00000173547
|
lctla
|
lactase-like a |
| chr8_+_14058646 | 0.33 |
ENSDART00000080852
|
ugt5e1
|
UDP glucuronosyltransferase 5 family, polypeptide E1 |
| chr16_+_17763848 | 0.33 |
ENSDART00000149408
ENSDART00000148878 |
them4
|
thioesterase superfamily member 4 |
| chr14_-_9400552 | 0.33 |
ENSDART00000129485
ENSDART00000131341 |
tbx22
|
T-box 22 |
| chr5_+_30179010 | 0.33 |
ENSDART00000134624
|
adamts15a
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15a |
| chr10_+_15088534 | 0.32 |
ENSDART00000142865
|
si:ch211-95j8.3
|
si:ch211-95j8.3 |
| chr21_-_37951819 | 0.32 |
ENSDART00000139549
|
si:dkey-38k9.5
|
si:dkey-38k9.5 |
| chr23_-_14990865 | 0.32 |
ENSDART00000147799
|
ndrg3b
|
ndrg family member 3b |
| chr24_-_32522587 | 0.32 |
ENSDART00000048968
ENSDART00000143781 |
EIF1B
|
zgc:56676 |
| chr8_+_25302172 | 0.32 |
ENSDART00000046182
ENSDART00000145316 |
gstm.3
|
glutathione S-transferase mu tandem duplicate 3 |
| chr8_-_13029297 | 0.32 |
ENSDART00000144305
|
dennd2da
|
DENN/MADD domain containing 2Da |
| chr22_-_20720427 | 0.31 |
ENSDART00000105532
|
oaz1a
|
ornithine decarboxylase antizyme 1a |
| chr10_-_39154594 | 0.31 |
ENSDART00000148825
|
slc37a4b
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4b |
| chr9_+_13999620 | 0.31 |
ENSDART00000143229
|
cd28l
|
cd28-like molecule |
| chr21_+_11685009 | 0.31 |
ENSDART00000014668
|
pcsk1
|
proprotein convertase subtilisin/kexin type 1 |
| chr12_-_34035364 | 0.31 |
ENSDART00000087065
|
timp2a
|
TIMP metallopeptidase inhibitor 2a |
| chr18_+_8805575 | 0.30 |
ENSDART00000137098
|
impdh1a
|
IMP (inosine 5'-monophosphate) dehydrogenase 1a |
| chr6_-_52796212 | 0.30 |
ENSDART00000154133
|
rbpjl
|
recombination signal binding protein for immunoglobulin kappa J region-like |
| chr2_+_1486822 | 0.30 |
ENSDART00000132500
|
c8a
|
complement component 8, alpha polypeptide |
| chr1_-_45888608 | 0.30 |
ENSDART00000139219
|
pnpla6
|
patatin-like phospholipase domain containing 6 |
| chr6_-_39218609 | 0.30 |
ENSDART00000133305
|
os9
|
osteosarcoma amplified 9, endoplasmic reticulum lectin |
| chr3_-_11878490 | 0.30 |
ENSDART00000129961
|
coro7
|
coronin 7 |
| chr21_+_11684830 | 0.29 |
ENSDART00000147473
|
pcsk1
|
proprotein convertase subtilisin/kexin type 1 |
| chr8_-_9118958 | 0.29 |
ENSDART00000037922
|
slc6a8
|
solute carrier family 6 (neurotransmitter transporter), member 8 |
| chr4_+_74255904 | 0.29 |
ENSDART00000174385
|
ptprr
|
protein tyrosine phosphatase, receptor type, r |
| chr5_-_64823750 | 0.29 |
ENSDART00000140305
|
lix1
|
limb and CNS expressed 1 |
| chr18_+_20468157 | 0.29 |
ENSDART00000100665
ENSDART00000147867 ENSDART00000060302 ENSDART00000180370 |
ddb2
|
damage-specific DNA binding protein 2 |
| chr7_-_67214972 | 0.29 |
ENSDART00000156861
|
swap70a
|
switching B cell complex subunit SWAP70a |
| chr21_-_40015530 | 0.29 |
ENSDART00000084237
|
slc47a2.1
|
solute carrier family 47 (multidrug and toxin extrusion), member 2.1 |
| chr11_-_34522249 | 0.29 |
ENSDART00000158616
|
pik3ca
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha |
| chr21_-_25573064 | 0.29 |
ENSDART00000134310
|
CR388166.1
|
|
| chr15_+_28106498 | 0.29 |
ENSDART00000041707
|
unc119a
|
unc-119 homolog a (C. elegans) |
| chr6_-_49873020 | 0.28 |
ENSDART00000148511
|
gnas
|
GNAS complex locus |
| chr16_-_11779508 | 0.27 |
ENSDART00000136329
ENSDART00000060145 ENSDART00000141101 |
pafah1b3
|
platelet-activating factor acetylhydrolase, isoform Ib, gamma subunit |
| chr21_-_14815952 | 0.27 |
ENSDART00000134278
ENSDART00000067004 |
phpt1
|
phosphohistidine phosphatase 1 |
| chr19_-_41213718 | 0.27 |
ENSDART00000077121
|
pdk4
|
pyruvate dehydrogenase kinase, isozyme 4 |
| chr11_+_25064519 | 0.27 |
ENSDART00000016181
|
ndrg3a
|
ndrg family member 3a |
| chr13_-_33317323 | 0.27 |
ENSDART00000110295
ENSDART00000144848 ENSDART00000136701 |
tmem234
|
transmembrane protein 234 |
| chr6_-_40098641 | 0.26 |
ENSDART00000017402
|
ip6k2b
|
inositol hexakisphosphate kinase 2b |
| chr5_-_55752169 | 0.26 |
ENSDART00000097424
|
kcmf1
|
potassium channel modulatory factor 1 |
| chr9_-_15208129 | 0.26 |
ENSDART00000137043
ENSDART00000131512 |
pard3bb
|
par-3 family cell polarity regulator beta b |
| chr2_+_1487118 | 0.26 |
ENSDART00000147283
|
c8a
|
complement component 8, alpha polypeptide |
| chr19_+_47783137 | 0.26 |
ENSDART00000024777
ENSDART00000158979 |
c19h1orf109
|
c19h1orf109 homolog (H. sapiens) |
| chr25_-_5119162 | 0.26 |
ENSDART00000153961
|
shisal1b
|
shisa like 1b |
| chr13_-_32995324 | 0.26 |
ENSDART00000140542
ENSDART00000037740 |
kcnf1b
|
potassium voltage-gated channel, subfamily F, member 1b |
| chr2_+_243778 | 0.26 |
ENSDART00000182262
|
CABZ01085887.1
|
|
| chr4_-_16644708 | 0.26 |
ENSDART00000042307
|
sinhcaf
|
SIN3-HDAC complex associated factor |
| chr18_-_14691727 | 0.26 |
ENSDART00000010129
|
pdf
|
peptide deformylase, mitochondrial |
| chr13_+_36770738 | 0.26 |
ENSDART00000146696
|
atl1
|
atlastin GTPase 1 |
| chr4_+_4267451 | 0.26 |
ENSDART00000192069
|
ano2
|
anoctamin 2 |
| chr23_+_40460333 | 0.26 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr14_+_29200772 | 0.25 |
ENSDART00000166608
|
TENM2
|
si:dkey-34l15.2 |
| chr14_+_31739568 | 0.25 |
ENSDART00000183507
|
adgrg4a
|
adhesion G protein-coupled receptor G4a |
| chr16_-_41465542 | 0.25 |
ENSDART00000169116
ENSDART00000187446 |
cpne4a
|
copine IVa |
| chr6_-_7439490 | 0.25 |
ENSDART00000188825
|
fkbp11
|
FK506 binding protein 11 |
| chr5_-_37900350 | 0.25 |
ENSDART00000084839
ENSDART00000084841 ENSDART00000133437 |
tmprss13b
|
transmembrane protease, serine 13b |
| chr24_+_20927135 | 0.25 |
ENSDART00000144883
ENSDART00000131829 |
fam162a
|
family with sequence similarity 162, member A |
| chr10_-_10018120 | 0.25 |
ENSDART00000132375
|
strbp
|
spermatid perinuclear RNA binding protein |
| chr1_-_28089557 | 0.25 |
ENSDART00000161024
ENSDART00000167875 |
snapc3
|
small nuclear RNA activating complex, polypeptide 3 |
| chr20_+_7084154 | 0.25 |
ENSDART00000136448
|
ftr85
|
finTRIM family, member 85 |
| chr21_+_9628854 | 0.25 |
ENSDART00000161753
ENSDART00000160711 |
mapk10
|
mitogen-activated protein kinase 10 |
| chr8_+_51050554 | 0.24 |
ENSDART00000166249
|
DISP3
|
si:dkey-32e23.6 |
| chr10_-_22127942 | 0.24 |
ENSDART00000133374
|
ponzr2
|
plac8 onzin related protein 2 |
| chr21_-_37733571 | 0.24 |
ENSDART00000176214
|
mpp1
|
membrane protein, palmitoylated 1 |
| chr15_-_16070731 | 0.24 |
ENSDART00000122099
|
dynll2a
|
dynein, light chain, LC8-type 2a |
| chr2_-_17044959 | 0.24 |
ENSDART00000090260
|
clcn2a
|
chloride channel, voltage-sensitive 2a |
| chr8_+_3379815 | 0.24 |
ENSDART00000155995
|
FUT9 (1 of many)
|
zgc:136963 |
| chr5_-_68927728 | 0.24 |
ENSDART00000132838
|
ank1a
|
ankyrin 1, erythrocytic a |
| chr21_-_14803366 | 0.24 |
ENSDART00000190872
|
si:dkey-11o18.5
|
si:dkey-11o18.5 |
| chr16_-_22225295 | 0.24 |
ENSDART00000163519
|
LO017682.1
|
|
| chr2_+_50477779 | 0.23 |
ENSDART00000122716
|
CABZ01067973.1
|
|
| chr11_+_30314885 | 0.23 |
ENSDART00000187418
ENSDART00000123244 |
ugt1b2
|
UDP glucuronosyltransferase 1 family, polypeptide B2 |
| chr24_+_22039964 | 0.23 |
ENSDART00000081220
|
ankrd33ba
|
ankyrin repeat domain 33ba |
| chr16_+_21801277 | 0.23 |
ENSDART00000088407
|
trim108
|
tripartite motif containing 108 |
| chr23_+_33907899 | 0.23 |
ENSDART00000159445
|
cs
|
citrate synthase |
| chr13_-_15702672 | 0.23 |
ENSDART00000144445
ENSDART00000168950 |
ckba
|
creatine kinase, brain a |
| chr23_-_27701361 | 0.23 |
ENSDART00000186688
ENSDART00000183985 |
dnajc22
|
DnaJ (Hsp40) homolog, subfamily C, member 22 |
| chr12_-_6880694 | 0.23 |
ENSDART00000171846
|
pcdh15b
|
protocadherin-related 15b |
| chr11_+_34522554 | 0.22 |
ENSDART00000109833
|
zmat3
|
zinc finger, matrin-type 3 |
| chr8_+_20624510 | 0.22 |
ENSDART00000138604
|
nfic
|
nuclear factor I/C |
| chr11_+_42422638 | 0.22 |
ENSDART00000042599
ENSDART00000181175 |
dennd6aa
|
DENN/MADD domain containing 6Aa |
| chr20_+_66857 | 0.22 |
ENSDART00000114999
|
lrfn2b
|
leucine rich repeat and fibronectin type III domain containing 2b |
| chr19_+_29798064 | 0.21 |
ENSDART00000167803
ENSDART00000051804 |
marcksl1b
|
MARCKS-like 1b |
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx2.2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0019556 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.3 | 3.3 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.2 | 0.6 | GO:0097623 | potassium ion export(GO:0071435) potassium ion export across plasma membrane(GO:0097623) |
| 0.2 | 1.0 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.2 | 0.5 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.2 | 0.6 | GO:0006530 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.1 | 0.6 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 0.3 | GO:0090278 | negative regulation of peptide secretion(GO:0002792) negative regulation of peptide hormone secretion(GO:0090278) |
| 0.1 | 0.3 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 0.9 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 0.5 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.1 | 0.4 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.1 | 0.3 | GO:0035971 | peptidyl-histidine dephosphorylation(GO:0035971) |
| 0.1 | 0.3 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.1 | 0.4 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.1 | 0.4 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.1 | 0.2 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 | 0.3 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.1 | 0.2 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.1 | 0.2 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.1 | 0.2 | GO:0090199 | regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.1 | 0.2 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.1 | 0.2 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.1 | 0.3 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.1 | 1.2 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.7 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.3 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.5 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.0 | 0.7 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.0 | 0.2 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.0 | 0.3 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.5 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.9 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.4 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.6 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.4 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.3 | GO:1900186 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.3 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.3 | GO:0055062 | phosphate ion homeostasis(GO:0055062) trivalent inorganic anion homeostasis(GO:0072506) |
| 0.0 | 0.6 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.3 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.5 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.0 | 0.5 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.3 | GO:0061615 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.3 | GO:0006177 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.0 | 0.1 | GO:0046351 | sucrose metabolic process(GO:0005985) sucrose biosynthetic process(GO:0005986) disaccharide biosynthetic process(GO:0046351) |
| 0.0 | 0.1 | GO:0072047 | proximal/distal pattern formation involved in nephron development(GO:0072047) distal tubule morphogenesis(GO:0072156) proximal/distal pattern formation involved in pronephric nephron development(GO:0072196) |
| 0.0 | 0.2 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.0 | 0.1 | GO:0046125 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.0 | 0.2 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.7 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.0 | 0.3 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 0.1 | GO:0021698 | cerebellar Purkinje cell layer structural organization(GO:0021693) cerebellar cortex structural organization(GO:0021698) |
| 0.0 | 0.4 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.8 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.7 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.2 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 4.4 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 1.5 | GO:0048841 | regulation of axon extension involved in axon guidance(GO:0048841) |
| 0.0 | 0.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.5 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.1 | GO:0006972 | hyperosmotic response(GO:0006972) |
| 0.0 | 0.1 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.3 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.0 | 0.1 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.0 | 0.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.2 | GO:0042396 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 0.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.3 | GO:0090316 | positive regulation of intracellular protein transport(GO:0090316) |
| 0.0 | 0.2 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.6 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.0 | GO:2000378 | negative regulation of gliogenesis(GO:0014014) positive regulation of DNA binding(GO:0043388) negative regulation of reactive oxygen species metabolic process(GO:2000378) |
| 0.0 | 0.1 | GO:0086010 | membrane depolarization(GO:0051899) membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.0 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.7 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.0 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.1 | GO:0034695 | response to prostaglandin E(GO:0034695) cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.0 | 0.0 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.0 | 0.1 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.1 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.0 | 0.4 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.3 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.1 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.0 | 0.6 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.1 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 | 0.3 | GO:0009411 | response to UV(GO:0009411) |
| 0.0 | 0.0 | GO:0042543 | protein N-linked glycosylation via arginine(GO:0042543) |
| 0.0 | 0.1 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.4 | GO:0048920 | posterior lateral line neuromast primordium migration(GO:0048920) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 1.7 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.1 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.2 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.6 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 1.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 0.7 | GO:0071914 | prominosome(GO:0071914) |
| 0.1 | 0.2 | GO:0019185 | snRNA-activating protein complex(GO:0019185) |
| 0.1 | 1.3 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 0.5 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 0.3 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 0.9 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.2 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.8 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.4 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.3 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.8 | GO:0032590 | neuron projection membrane(GO:0032589) dendrite membrane(GO:0032590) |
| 0.0 | 0.3 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.5 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 1.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.0 | 0.1 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.5 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.1 | GO:1990923 | PET complex(GO:1990923) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.2 | 0.9 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.2 | 0.7 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.6 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.1 | 0.6 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 0.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.3 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.4 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.1 | 0.3 | GO:0101006 | protein histidine phosphatase activity(GO:0101006) |
| 0.1 | 0.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 0.2 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 1.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.3 | GO:0051430 | mu-type opioid receptor binding(GO:0031852) dopamine receptor binding(GO:0050780) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 0.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.3 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.8 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.6 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.8 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.4 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.7 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.5 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.2 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.0 | 0.3 | GO:0001032 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 3 promoter sequence-specific DNA binding(GO:0001006) RNA polymerase III regulatory region DNA binding(GO:0001016) RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.0 | 0.5 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 1.0 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.9 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.4 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 3.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 1.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.3 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.3 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.3 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.2 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.2 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.3 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.3 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.3 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.7 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.4 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.1 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 1.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.8 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.6 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 0.1 | GO:0004133 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.0 | 0.5 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.4 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.1 | GO:0052726 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) |
| 0.0 | 0.1 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.0 | 3.2 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 0.2 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.1 | GO:0002061 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.0 | 3.0 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.3 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.3 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.8 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.3 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.4 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.2 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.0 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.8 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.7 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.1 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.1 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 0.4 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.2 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 0.6 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.1 | 0.3 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.6 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 1.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.3 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.3 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 2.0 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.3 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.8 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |