Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
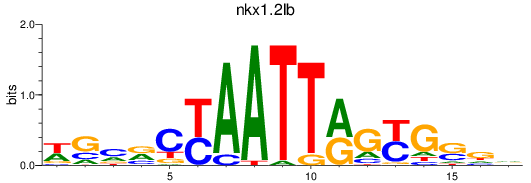
Results for nkx1.2lb
Z-value: 0.49
Transcription factors associated with nkx1.2lb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx1.2lb
|
ENSDARG00000099427 | NK1 transcription factor related 2-like,b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nkx1.2lb | dr11_v1_chr14_+_15064870_15064870 | 0.09 | 7.1e-01 | Click! |
Activity profile of nkx1.2lb motif
Sorted Z-values of nkx1.2lb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_306036 | 1.80 |
ENSDART00000103659
|
msgn1
|
mesogenin 1 |
| chr7_-_64971839 | 0.96 |
ENSDART00000164682
|
sinhcafl
|
SIN3-HDAC complex associated factor, like |
| chr17_-_15189397 | 0.95 |
ENSDART00000133710
ENSDART00000110507 |
wdhd1
|
WD repeat and HMG-box DNA binding protein 1 |
| chr8_-_16697615 | 0.93 |
ENSDART00000187929
|
rpe65b
|
retinal pigment epithelium-specific protein 65b |
| chr12_-_43685802 | 0.87 |
ENSDART00000170723
|
zgc:112964
|
zgc:112964 |
| chr8_-_50888806 | 0.84 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr8_-_20243389 | 0.82 |
ENSDART00000184904
|
acer1
|
alkaline ceramidase 1 |
| chr8_-_39822917 | 0.79 |
ENSDART00000067843
|
zgc:162025
|
zgc:162025 |
| chr4_+_6032640 | 0.78 |
ENSDART00000157487
|
tfec
|
transcription factor EC |
| chr20_+_36812368 | 0.77 |
ENSDART00000062931
|
abracl
|
ABRA C-terminal like |
| chr21_-_31013817 | 0.65 |
ENSDART00000065504
|
ncbp3
|
nuclear cap binding subunit 3 |
| chr17_+_16429826 | 0.65 |
ENSDART00000136078
|
efcab11
|
EF-hand calcium binding domain 11 |
| chr23_+_29358188 | 0.64 |
ENSDART00000189242
|
tardbpl
|
TAR DNA binding protein, like |
| chr5_-_22052852 | 0.62 |
ENSDART00000002938
|
mtmr8
|
myotubularin related protein 8 |
| chr13_-_9213207 | 0.61 |
ENSDART00000139861
ENSDART00000140524 |
HTRA2 (1 of many)
|
si:dkey-33c12.11 |
| chr11_+_16153207 | 0.58 |
ENSDART00000192356
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr7_+_24881680 | 0.58 |
ENSDART00000058843
|
krcp
|
kelch repeat-containing protein |
| chr17_+_24821627 | 0.54 |
ENSDART00000112389
|
wdr43
|
WD repeat domain 43 |
| chr2_+_6253246 | 0.54 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr11_-_16152400 | 0.54 |
ENSDART00000123665
|
arpc4l
|
actin related protein 2/3 complex, subunit 4, like |
| chr21_-_15200556 | 0.53 |
ENSDART00000141809
|
sfswap
|
splicing factor SWAP |
| chr21_-_26071773 | 0.53 |
ENSDART00000141382
|
rab34b
|
RAB34, member RAS oncogene family b |
| chr5_-_51619742 | 0.52 |
ENSDART00000188537
|
otpb
|
orthopedia homeobox b |
| chr18_+_9171778 | 0.52 |
ENSDART00000101192
|
sema3d
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D |
| chr11_+_16152316 | 0.51 |
ENSDART00000081054
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr10_+_39212898 | 0.51 |
ENSDART00000159501
|
stt3a
|
STT3A, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr15_-_25518084 | 0.50 |
ENSDART00000158594
|
hif1al
|
hypoxia-inducible factor 1, alpha subunit, like |
| chr23_-_24263474 | 0.49 |
ENSDART00000160312
|
hspb7
|
heat shock protein family, member 7 (cardiovascular) |
| chr13_-_9159852 | 0.47 |
ENSDART00000170185
ENSDART00000158697 ENSDART00000143393 ENSDART00000164973 ENSDART00000159910 |
HTRA2 (1 of many)
|
si:dkey-112g5.16 |
| chr21_+_4410733 | 0.47 |
ENSDART00000136831
ENSDART00000131612 |
HTRA2 (1 of many)
|
si:dkey-84o3.7 |
| chr8_+_18011522 | 0.45 |
ENSDART00000136756
|
ssbp3b
|
single stranded DNA binding protein 3b |
| chr6_-_7720332 | 0.45 |
ENSDART00000135945
|
rpsa
|
ribosomal protein SA |
| chr2_+_10006839 | 0.44 |
ENSDART00000160304
|
slc35a3b
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3b |
| chr21_+_4328348 | 0.44 |
ENSDART00000131658
ENSDART00000143799 |
HTRA2 (1 of many)
|
si:dkey-84o3.7 |
| chr13_-_9236905 | 0.44 |
ENSDART00000142597
ENSDART00000147176 ENSDART00000181670 |
HTRA2 (1 of many)
si:dkey-112g5.12
|
si:dkey-33c12.12 si:dkey-112g5.12 |
| chr23_+_43255328 | 0.44 |
ENSDART00000102712
|
tgm2a
|
transglutaminase 2, C polypeptide A |
| chr10_-_34002185 | 0.43 |
ENSDART00000046599
|
zar1l
|
zygote arrest 1-like |
| chr9_+_43799829 | 0.41 |
ENSDART00000186240
|
ube2e3
|
ubiquitin-conjugating enzyme E2E 3 (UBC4/5 homolog, yeast) |
| chr13_-_9045879 | 0.40 |
ENSDART00000155463
ENSDART00000140041 ENSDART00000137454 |
si:dkey-112g5.11
|
si:dkey-112g5.11 |
| chr5_-_33298352 | 0.40 |
ENSDART00000015076
|
zgc:103692
|
zgc:103692 |
| chr21_+_4348600 | 0.38 |
ENSDART00000138463
|
HTRA2 (1 of many)
|
si:dkey-84o3.2 |
| chr13_+_9521629 | 0.37 |
ENSDART00000149870
ENSDART00000137666 |
si:dkey-19p15.4
|
si:dkey-19p15.4 |
| chr13_+_9468535 | 0.37 |
ENSDART00000135088
ENSDART00000164270 ENSDART00000099619 ENSDART00000164656 |
HTRA2 (1 of many)
|
si:dkey-265c15.6 |
| chr1_-_48933 | 0.35 |
ENSDART00000171162
|
ildr1a
|
immunoglobulin-like domain containing receptor 1a |
| chr6_-_47840465 | 0.35 |
ENSDART00000141986
|
lrig2
|
leucine-rich repeats and immunoglobulin-like domains 2 |
| chr17_+_21902058 | 0.34 |
ENSDART00000142178
|
plekha1a
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1a |
| chr11_+_12811906 | 0.34 |
ENSDART00000123445
|
rtel1
|
regulator of telomere elongation helicase 1 |
| chr1_-_8140763 | 0.34 |
ENSDART00000091508
|
FAM83G
|
si:dkeyp-9d4.4 |
| chr23_-_31913069 | 0.33 |
ENSDART00000135526
|
mtfr2
|
mitochondrial fission regulator 2 |
| chr13_+_35955562 | 0.33 |
ENSDART00000137377
|
si:ch211-67f13.7
|
si:ch211-67f13.7 |
| chr13_-_9257728 | 0.33 |
ENSDART00000145985
ENSDART00000169991 ENSDART00000132764 |
HTRA2 (1 of many)
|
zgc:173425 |
| chr25_+_20272145 | 0.33 |
ENSDART00000109605
|
si:dkey-219c3.2
|
si:dkey-219c3.2 |
| chr21_+_4368735 | 0.32 |
ENSDART00000182817
|
CT027677.1
|
|
| chr6_+_52877147 | 0.32 |
ENSDART00000174370
|
or137-8
|
odorant receptor, family H, subfamily 137, member 8 |
| chr2_-_24317240 | 0.32 |
ENSDART00000078975
|
trnau1apb
|
tRNA selenocysteine 1 associated protein 1b |
| chr13_-_36798204 | 0.32 |
ENSDART00000012357
|
sav1
|
salvador family WW domain containing protein 1 |
| chr4_-_29753895 | 0.32 |
ENSDART00000150742
|
si:dkey-191j3.1
|
si:dkey-191j3.1 |
| chr21_+_4429001 | 0.31 |
ENSDART00000134520
|
HTRA2 (1 of many)
|
si:dkey-84o3.6 |
| chr13_-_9070754 | 0.31 |
ENSDART00000143783
ENSDART00000102121 ENSDART00000140820 ENSDART00000184210 |
si:dkey-112g5.12
|
si:dkey-112g5.12 |
| chr13_-_9111927 | 0.30 |
ENSDART00000133815
ENSDART00000109783 ENSDART00000142540 ENSDART00000192613 |
HTRA2 (1 of many)
si:dkey-112g5.12
|
si:dkey-19p15.4 si:dkey-112g5.12 |
| chr9_+_35015747 | 0.30 |
ENSDART00000140110
|
gabpa
|
GA binding protein transcription factor, alpha subunit |
| chr6_+_7421898 | 0.30 |
ENSDART00000043946
|
ccdc65
|
coiled-coil domain containing 65 |
| chr13_-_9184132 | 0.29 |
ENSDART00000135289
ENSDART00000136309 ENSDART00000132630 |
HTRA2 (1 of many)
|
si:dkey-33c12.10 |
| chr20_-_37813863 | 0.29 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr2_+_25315591 | 0.28 |
ENSDART00000161386
|
fndc3ba
|
fibronectin type III domain containing 3Ba |
| chr14_-_215051 | 0.27 |
ENSDART00000054822
|
nkx3.2
|
NK3 homeobox 2 |
| chr4_+_63790028 | 0.27 |
ENSDART00000160964
|
si:dkey-92j16.2
|
si:dkey-92j16.2 |
| chr21_+_16145980 | 0.27 |
ENSDART00000135119
|
qrfpr4
|
pyroglutamylated RFamide peptide receptor 4 |
| chr12_-_4249000 | 0.27 |
ENSDART00000059298
|
zgc:92313
|
zgc:92313 |
| chr6_+_23026714 | 0.26 |
ENSDART00000124948
|
srp68
|
signal recognition particle 68 |
| chr21_-_21570134 | 0.26 |
ENSDART00000174388
|
or133-4
|
odorant receptor, family H, subfamily 133, member 4 |
| chr4_+_11458078 | 0.25 |
ENSDART00000037600
|
ankrd16
|
ankyrin repeat domain 16 |
| chr13_+_411607 | 0.25 |
ENSDART00000186483
|
CU570800.3
|
|
| chr12_-_19119176 | 0.25 |
ENSDART00000149180
|
aco2
|
aconitase 2, mitochondrial |
| chr13_+_9496748 | 0.25 |
ENSDART00000147050
ENSDART00000102120 |
CR848040.4
|
|
| chr6_-_12275836 | 0.24 |
ENSDART00000189980
|
pkp4
|
plakophilin 4 |
| chr7_-_40630698 | 0.23 |
ENSDART00000134547
|
ube3c
|
ubiquitin protein ligase E3C |
| chr25_-_27819838 | 0.23 |
ENSDART00000067106
|
lmod2a
|
leiomodin 2 (cardiac) a |
| chr19_-_18313303 | 0.22 |
ENSDART00000164644
ENSDART00000167480 ENSDART00000163104 |
si:dkey-208k4.2
|
si:dkey-208k4.2 |
| chr10_+_27021164 | 0.21 |
ENSDART00000183538
|
BX248510.12
|
|
| chr18_+_13236255 | 0.20 |
ENSDART00000091560
|
tldc1
|
TBC/LysM-associated domain containing 1 |
| chr24_-_32582378 | 0.19 |
ENSDART00000066590
|
rdh12l
|
retinol dehydrogenase 12, like |
| chr6_+_31368071 | 0.19 |
ENSDART00000038990
|
jak1
|
Janus kinase 1 |
| chr22_-_17458070 | 0.19 |
ENSDART00000139658
|
si:ch211-197g15.10
|
si:ch211-197g15.10 |
| chr13_-_17723417 | 0.18 |
ENSDART00000183834
|
vdac2
|
voltage-dependent anion channel 2 |
| chr8_-_29658171 | 0.18 |
ENSDART00000147755
|
mpeg1.3
|
macrophage expressed 1, tandem duplicate 3 |
| chr19_-_10810006 | 0.16 |
ENSDART00000151157
|
si:dkey-3n22.9
|
si:dkey-3n22.9 |
| chr8_-_17822259 | 0.16 |
ENSDART00000063587
|
st6galnac5b
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5b |
| chr20_-_14114078 | 0.16 |
ENSDART00000168434
ENSDART00000104032 |
si:ch211-223m11.2
|
si:ch211-223m11.2 |
| chr7_+_34453185 | 0.15 |
ENSDART00000173875
ENSDART00000173921 ENSDART00000173995 |
si:cabz01009626.1
|
si:cabz01009626.1 |
| chr19_+_26718074 | 0.15 |
ENSDART00000134455
|
zgc:100906
|
zgc:100906 |
| chr16_+_10318893 | 0.15 |
ENSDART00000055380
|
tubb5
|
tubulin, beta 5 |
| chr2_+_19522082 | 0.14 |
ENSDART00000146098
|
pimr49
|
Pim proto-oncogene, serine/threonine kinase, related 49 |
| chr11_+_31864921 | 0.14 |
ENSDART00000180252
|
diaph3
|
diaphanous-related formin 3 |
| chr13_-_6279684 | 0.14 |
ENSDART00000092105
|
agpat5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 (lysophosphatidic acid acyltransferase, epsilon) |
| chr7_-_59123066 | 0.14 |
ENSDART00000175438
|
dennd4c
|
DENN/MADD domain containing 4C |
| chr17_-_53329704 | 0.14 |
ENSDART00000193895
|
exd1
|
exonuclease 3'-5' domain containing 1 |
| chr2_+_10007113 | 0.13 |
ENSDART00000155213
|
slc35a3b
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3b |
| chr2_+_19578079 | 0.13 |
ENSDART00000144413
|
pimr50
|
Pim proto-oncogene, serine/threonine kinase, related 50 |
| chr13_-_30142087 | 0.11 |
ENSDART00000110157
|
tysnd1
|
trypsin domain containing 1 |
| chr1_-_44812014 | 0.11 |
ENSDART00000139044
|
si:dkey-9i23.8
|
si:dkey-9i23.8 |
| chr4_-_9891874 | 0.11 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr20_+_14114258 | 0.08 |
ENSDART00000044937
|
kcns3b
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3b |
| chr13_-_36663358 | 0.07 |
ENSDART00000085319
|
sos2
|
son of sevenless homolog 2 (Drosophila) |
| chr7_+_26173751 | 0.07 |
ENSDART00000065131
|
si:ch211-196f2.7
|
si:ch211-196f2.7 |
| chr4_+_73215536 | 0.06 |
ENSDART00000174290
|
LO018260.2
|
Danio rerio protein NLRC3-like (LOC101883187), mRNA. |
| chr25_-_7705487 | 0.06 |
ENSDART00000128099
|
prdm11
|
PR domain containing 11 |
| chr3_-_40836081 | 0.06 |
ENSDART00000143135
|
wipi2
|
WD repeat domain, phosphoinositide interacting 2 |
| chr25_+_469855 | 0.06 |
ENSDART00000104717
|
rsl24d1
|
ribosomal L24 domain containing 1 |
| chr25_+_36312459 | 0.05 |
ENSDART00000182484
|
CR354435.5
|
|
| chr8_-_4031121 | 0.04 |
ENSDART00000169474
ENSDART00000163754 |
mtmr3
|
myotubularin related protein 3 |
| chr14_+_45306073 | 0.03 |
ENSDART00000034606
ENSDART00000173301 |
sfxn5b
|
sideroflexin 5b |
| chr12_+_3078221 | 0.01 |
ENSDART00000148835
ENSDART00000149427 |
sgca
|
sarcoglycan, alpha |
| chr1_-_50791280 | 0.00 |
ENSDART00000181224
|
CABZ01031870.1
|
|
| chr1_+_55758257 | 0.00 |
ENSDART00000139312
|
tecrb
|
trans-2,3-enoyl-CoA reductase b |
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx1.2lb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0048341 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) |
| 0.2 | 0.9 | GO:0016116 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 0.1 | 0.8 | GO:0097065 | anterior head development(GO:0097065) |
| 0.1 | 0.7 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 | 0.5 | GO:0007508 | larval development(GO:0002164) larval heart development(GO:0007508) |
| 0.1 | 0.5 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.5 | GO:0021767 | mammillary body development(GO:0021767) |
| 0.1 | 0.8 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.1 | 0.8 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.1 | 0.5 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 0.1 | 0.5 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.1 | 0.9 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.6 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.3 | GO:0032205 | negative regulation of telomere maintenance(GO:0032205) |
| 0.0 | 0.3 | GO:0003352 | regulation of cilium movement(GO:0003352) |
| 0.0 | 0.4 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 1.1 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.0 | 0.8 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.5 | GO:0043687 | protein N-linked glycosylation via asparagine(GO:0018279) post-translational protein modification(GO:0043687) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.5 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.6 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.4 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.2 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.6 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.1 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 0.2 | GO:0033077 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.0 | 0.3 | GO:0006376 | mRNA splice site selection(GO:0006376) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.3 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.1 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.9 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 0.5 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.3 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.3 | GO:0005858 | axonemal dynein complex(GO:0005858) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0052885 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.1 | 0.8 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.5 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 0.3 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 0.2 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 0.1 | 0.8 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 0.7 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.6 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 0.9 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.3 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.7 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.2 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 1.1 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.4 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.3 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 0.3 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.8 | PID CMYB PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |