Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
Results for nfkb1_rela
Z-value: 0.94
Transcription factors associated with nfkb1_rela
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
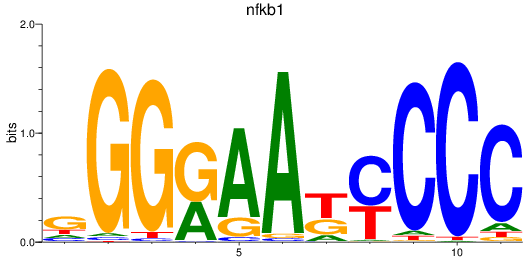
nfkb1
|
ENSDARG00000105261 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
|
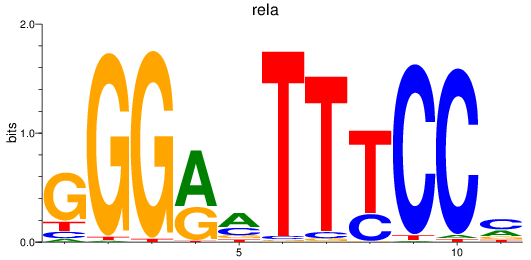
rela
|
ENSDARG00000098696 | v-rel avian reticuloendotheliosis viral oncogene homolog A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nfkb1 | dr11_v1_chr14_-_49859747_49859823 | 0.35 | 1.4e-01 | Click! |
| rela | dr11_v1_chr7_-_804515_804515 | -0.34 | 1.6e-01 | Click! |
Activity profile of nfkb1_rela motif
Sorted Z-values of nfkb1_rela motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_50175069 | 2.93 |
ENSDART00000192979
|
lim2.5
|
lens intrinsic membrane protein 2.5 |
| chr3_-_28120092 | 2.83 |
ENSDART00000151143
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr23_+_45579497 | 2.60 |
ENSDART00000110381
|
egr4
|
early growth response 4 |
| chr2_+_42191592 | 2.27 |
ENSDART00000144716
|
cavin4a
|
caveolae associated protein 4a |
| chr21_+_28958471 | 2.22 |
ENSDART00000144331
ENSDART00000005929 |
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr19_+_19412692 | 2.21 |
ENSDART00000113580
|
wu:fc38h03
|
wu:fc38h03 |
| chr3_-_32170850 | 2.21 |
ENSDART00000055307
ENSDART00000157366 |
tnnt1
|
troponin T type 1 (skeletal, slow) |
| chr3_+_11568523 | 2.10 |
ENSDART00000179668
|
CABZ01087224.1
|
|
| chr25_+_37397031 | 1.95 |
ENSDART00000193643
ENSDART00000169132 |
slc1a2b
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2b |
| chr1_-_30039331 | 1.74 |
ENSDART00000086935
ENSDART00000143800 |
MARCH4 (1 of many)
|
zgc:153256 |
| chr15_-_24869826 | 1.71 |
ENSDART00000127047
|
tusc5a
|
tumor suppressor candidate 5a |
| chr6_-_10305918 | 1.69 |
ENSDART00000090994
|
scn1lab
|
sodium channel, voltage-gated, type I like, alpha b |
| chr10_-_39011514 | 1.63 |
ENSDART00000075123
|
pcp4a
|
Purkinje cell protein 4a |
| chr6_-_607063 | 1.61 |
ENSDART00000189900
|
lgals2b
|
lectin, galactoside-binding, soluble, 2b |
| chr3_+_46459540 | 1.61 |
ENSDART00000188150
|
si:ch211-66e2.5
|
si:ch211-66e2.5 |
| chr18_-_5527050 | 1.60 |
ENSDART00000145400
ENSDART00000132498 ENSDART00000146209 |
zgc:153317
|
zgc:153317 |
| chr1_-_45888608 | 1.59 |
ENSDART00000139219
|
pnpla6
|
patatin-like phospholipase domain containing 6 |
| chr22_+_14051245 | 1.57 |
ENSDART00000043711
ENSDART00000164259 |
aox6
|
aldehyde oxidase 6 |
| chr13_-_9318891 | 1.56 |
ENSDART00000137364
|
si:dkey-33c12.3
|
si:dkey-33c12.3 |
| chr14_-_49859747 | 1.55 |
ENSDART00000169456
ENSDART00000164967 |
nfkb1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
| chr6_+_2195625 | 1.52 |
ENSDART00000155659
|
acvr1bb
|
activin A receptor type 1Bb |
| chr1_+_42225060 | 1.51 |
ENSDART00000138740
ENSDART00000101306 |
ctnna2
|
catenin (cadherin-associated protein), alpha 2 |
| chr6_+_48664275 | 1.51 |
ENSDART00000161184
|
FAM19A3
|
si:dkey-238f9.1 |
| chr14_-_4121052 | 1.49 |
ENSDART00000167074
|
irf2
|
interferon regulatory factor 2 |
| chr4_+_18806251 | 1.45 |
ENSDART00000138662
|
slc26a3.2
|
solute carrier family 26 (anion exchanger), member 3, tandem duplicate 2 |
| chr1_+_32528097 | 1.43 |
ENSDART00000128317
|
nlgn4a
|
neuroligin 4a |
| chr14_-_31080183 | 1.42 |
ENSDART00000173282
|
mbnl3
|
muscleblind-like splicing regulator 3 |
| chr20_+_28434196 | 1.37 |
ENSDART00000034245
|
dpf3
|
D4, zinc and double PHD fingers, family 3 |
| chr20_+_16881883 | 1.34 |
ENSDART00000130107
|
nfkbiaa
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha a |
| chr20_+_33981946 | 1.33 |
ENSDART00000131775
|
si:dkey-51e6.1
|
si:dkey-51e6.1 |
| chr14_+_3038473 | 1.33 |
ENSDART00000026021
ENSDART00000150000 |
cd74a
|
CD74 molecule, major histocompatibility complex, class II invariant chain a |
| chr13_-_22843562 | 1.32 |
ENSDART00000142738
|
pbld
|
phenazine biosynthesis like protein domain containing |
| chr1_+_16397063 | 1.30 |
ENSDART00000159794
|
micu3a
|
mitochondrial calcium uptake family, member 3a |
| chr2_-_51772438 | 1.29 |
ENSDART00000170241
|
BX908782.2
|
Danio rerio three-finger protein 5 (LOC100003647), mRNA. |
| chr22_-_38137908 | 1.28 |
ENSDART00000104528
|
tm4sf4
|
transmembrane 4 L six family member 4 |
| chr17_-_51195651 | 1.26 |
ENSDART00000191205
ENSDART00000088185 |
paplna
|
papilin a, proteoglycan-like sulfated glycoprotein |
| chr16_+_1803462 | 1.25 |
ENSDART00000183974
|
GRIK2
|
glutamate ionotropic receptor kainate type subunit 2 |
| chr8_-_46572298 | 1.25 |
ENSDART00000030470
|
sult1st3
|
sulfotransferase family 1, cytosolic sulfotransferase 3 |
| chr23_-_32162810 | 1.24 |
ENSDART00000155905
|
nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr1_+_51896147 | 1.23 |
ENSDART00000167139
|
nfixa
|
nuclear factor I/Xa |
| chr22_+_110158 | 1.23 |
ENSDART00000143698
|
prkar2ab
|
protein kinase, cAMP-dependent, regulatory, type II, alpha, B |
| chr1_+_16396870 | 1.21 |
ENSDART00000189245
ENSDART00000113266 |
micu3a
|
mitochondrial calcium uptake family, member 3a |
| chr4_+_3389866 | 1.21 |
ENSDART00000153834
|
sypl1
|
synaptophysin-like 1 |
| chr21_+_25226558 | 1.19 |
ENSDART00000168480
|
sycn.2
|
syncollin, tandem duplicate 2 |
| chr13_-_2010191 | 1.19 |
ENSDART00000161021
ENSDART00000124134 |
gfral
|
GDNF family receptor alpha like |
| chr4_-_149334 | 1.18 |
ENSDART00000163280
|
tbk1
|
TANK-binding kinase 1 |
| chr25_-_27843066 | 1.16 |
ENSDART00000179684
ENSDART00000186000 ENSDART00000190065 |
asb15a
|
ankyrin repeat and SOCS box containing 15a |
| chr14_-_4120636 | 1.16 |
ENSDART00000059230
|
irf2
|
interferon regulatory factor 2 |
| chr25_+_5821910 | 1.14 |
ENSDART00000161035
|
C12orf75
|
chromosome 12 open reading frame 75 |
| chr7_+_39634873 | 1.14 |
ENSDART00000114774
|
ptpn5
|
protein tyrosine phosphatase, non-receptor type 5 |
| chr23_-_12423778 | 1.13 |
ENSDART00000124091
|
wfdc2
|
WAP four-disulfide core domain 2 |
| chr7_-_16204885 | 1.12 |
ENSDART00000171008
|
btr05
|
bloodthirsty-related gene family, member 5 |
| chr19_+_1606185 | 1.12 |
ENSDART00000092183
|
lrrc3b
|
leucine rich repeat containing 3B |
| chr24_+_26006730 | 1.10 |
ENSDART00000140384
ENSDART00000139184 |
ccl20b
|
chemokine (C-C motif) ligand 20b |
| chr11_-_32723851 | 1.05 |
ENSDART00000155592
|
pcdh17
|
protocadherin 17 |
| chr14_-_7888748 | 1.04 |
ENSDART00000166293
|
ppp3cb
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr12_+_48634927 | 1.04 |
ENSDART00000168441
|
zgc:165653
|
zgc:165653 |
| chr14_-_30967284 | 1.04 |
ENSDART00000149435
|
il2rgb
|
interleukin 2 receptor, gamma b |
| chr14_-_30642819 | 1.03 |
ENSDART00000078154
|
npas4a
|
neuronal PAS domain protein 4a |
| chr13_+_27316934 | 1.01 |
ENSDART00000164533
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr22_-_20011476 | 1.01 |
ENSDART00000093312
ENSDART00000093310 |
celf5a
|
cugbp, Elav-like family member 5a |
| chr2_+_45191049 | 1.01 |
ENSDART00000165392
|
ccl20a.3
|
chemokine (C-C motif) ligand 20a, duplicate 3 |
| chr19_-_44801918 | 0.98 |
ENSDART00000108507
|
CSMD3 (1 of many)
|
si:ch211-233f16.1 |
| chr1_+_12135129 | 0.97 |
ENSDART00000126020
|
spink4
|
serine peptidase inhibitor, Kazal type 4 |
| chr12_+_7445595 | 0.96 |
ENSDART00000103536
ENSDART00000152524 |
phyhiplb
|
phytanoyl-CoA 2-hydroxylase interacting protein-like b |
| chr17_+_20205185 | 0.96 |
ENSDART00000167007
|
gnrh3
|
gonadotropin-releasing hormone 3 |
| chr12_+_26471712 | 0.95 |
ENSDART00000162115
|
ndel1a
|
nudE neurodevelopment protein 1-like 1a |
| chr16_-_27749172 | 0.91 |
ENSDART00000145198
|
steap4
|
STEAP family member 4 |
| chr12_+_49100365 | 0.90 |
ENSDART00000171905
|
CABZ01075125.1
|
|
| chr21_-_435466 | 0.89 |
ENSDART00000110297
|
klf4
|
Kruppel-like factor 4 |
| chr20_+_22045089 | 0.88 |
ENSDART00000063564
ENSDART00000187013 ENSDART00000161552 ENSDART00000174478 ENSDART00000063568 ENSDART00000152247 |
nmu
|
neuromedin U |
| chr16_-_12984631 | 0.87 |
ENSDART00000184863
|
cacng7b
|
calcium channel, voltage-dependent, gamma subunit 7b |
| chr13_+_27316632 | 0.87 |
ENSDART00000016121
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr1_+_54677173 | 0.86 |
ENSDART00000114705
|
gprc5bb
|
G protein-coupled receptor, class C, group 5, member Bb |
| chr14_+_36889893 | 0.86 |
ENSDART00000124159
|
si:ch211-132p1.3
|
si:ch211-132p1.3 |
| chr3_-_40976463 | 0.84 |
ENSDART00000128450
ENSDART00000018676 |
cyp3c1
|
cytochrome P450, family 3, subfamily c, polypeptide 1 |
| chr2_-_50966124 | 0.84 |
ENSDART00000170470
|
ghrhra
|
growth hormone releasing hormone receptor a |
| chr25_+_10458990 | 0.83 |
ENSDART00000130354
ENSDART00000044738 |
ric8a
|
RIC8 guanine nucleotide exchange factor A |
| chr17_+_1063988 | 0.81 |
ENSDART00000160400
|
gchfr
|
GTP cyclohydrolase I feedback regulator |
| chr16_-_16590780 | 0.80 |
ENSDART00000059841
|
si:ch211-257p13.3
|
si:ch211-257p13.3 |
| chr3_-_21242460 | 0.80 |
ENSDART00000007293
|
tcap
|
titin-cap (telethonin) |
| chr13_-_45795722 | 0.80 |
ENSDART00000141779
|
fndc5a
|
fibronectin type III domain containing 5a |
| chr4_-_25485214 | 0.78 |
ENSDART00000159941
|
pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr20_-_8321280 | 0.78 |
ENSDART00000083906
ENSDART00000186477 |
dab1a
|
Dab, reelin signal transducer, homolog 1a (Drosophila) |
| chr10_-_14536399 | 0.78 |
ENSDART00000186501
|
hdhd2
|
haloacid dehalogenase-like hydrolase domain containing 2 |
| chr19_-_11208782 | 0.78 |
ENSDART00000044426
ENSDART00000189754 |
si:dkey-240h12.4
|
si:dkey-240h12.4 |
| chr13_+_12739283 | 0.77 |
ENSDART00000102279
|
lingo2b
|
leucine rich repeat and Ig domain containing 2b |
| chr9_+_53707240 | 0.77 |
ENSDART00000171490
|
PCDH8
|
si:ch211-199f5.1 |
| chr14_+_45028062 | 0.76 |
ENSDART00000184717
ENSDART00000185481 |
ATP8A1
|
ATPase phospholipid transporting 8A1 |
| chr24_-_12689571 | 0.76 |
ENSDART00000015517
|
pdcd6
|
programmed cell death 6 |
| chr4_-_25485404 | 0.75 |
ENSDART00000041402
|
pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr14_+_36885524 | 0.75 |
ENSDART00000032547
|
lect2l
|
leukocyte cell-derived chemotaxin 2 like |
| chr9_+_993477 | 0.75 |
ENSDART00000182045
|
CABZ01066921.1
|
|
| chr2_-_57227542 | 0.75 |
ENSDART00000182675
ENSDART00000159480 |
btbd2b
|
BTB (POZ) domain containing 2b |
| chr9_-_6380653 | 0.74 |
ENSDART00000078523
|
ecrg4a
|
esophageal cancer related gene 4a |
| chr9_+_31752628 | 0.74 |
ENSDART00000060054
|
itgbl1
|
integrin, beta-like 1 |
| chr17_+_8175998 | 0.74 |
ENSDART00000131200
|
myct1b
|
myc target 1b |
| chr8_+_53269657 | 0.73 |
ENSDART00000184212
|
gnb1a
|
guanine nucleotide binding protein (G protein), beta polypeptide 1a |
| chr16_-_41004731 | 0.73 |
ENSDART00000102591
|
KCNV1
|
si:dkey-201i6.2 |
| chr4_+_13810811 | 0.72 |
ENSDART00000067168
|
pdzrn4
|
PDZ domain containing ring finger 4 |
| chr24_-_33703504 | 0.71 |
ENSDART00000079292
|
cavin4b
|
caveolae associated protein 4b |
| chr2_+_6963296 | 0.71 |
ENSDART00000147146
|
ddr2b
|
discoidin domain receptor tyrosine kinase 2b |
| chr25_-_35960229 | 0.70 |
ENSDART00000073434
|
snx20
|
sorting nexin 20 |
| chr14_+_49376011 | 0.69 |
ENSDART00000020961
|
tnip1
|
TNFAIP3 interacting protein 1 |
| chr21_-_43088991 | 0.68 |
ENSDART00000189683
|
si:ch73-68b22.2
|
si:ch73-68b22.2 |
| chr16_+_46148990 | 0.68 |
ENSDART00000083919
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr2_-_9857932 | 0.67 |
ENSDART00000154490
|
si:ch211-252f13.5
|
si:ch211-252f13.5 |
| chr14_+_1719367 | 0.67 |
ENSDART00000157696
|
trpc7b
|
transient receptor potential cation channel, subfamily C, member 7b |
| chr20_+_48100261 | 0.67 |
ENSDART00000158604
|
xkr5a
|
XK related 5a |
| chr12_-_26153101 | 0.66 |
ENSDART00000076051
|
opn4b
|
opsin 4b |
| chr25_-_1235457 | 0.65 |
ENSDART00000093093
|
coro2bb
|
coronin, actin binding protein, 2Bb |
| chr16_+_10346277 | 0.65 |
ENSDART00000081092
|
si:dkeyp-77h1.4
|
si:dkeyp-77h1.4 |
| chr14_-_2221877 | 0.64 |
ENSDART00000106704
|
pcdh2ab1
|
protocadherin 2 alpha b 1 |
| chr23_+_3616224 | 0.63 |
ENSDART00000190917
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr12_+_20641471 | 0.63 |
ENSDART00000133654
|
calcoco2
|
calcium binding and coiled-coil domain 2 |
| chr2_+_21356242 | 0.63 |
ENSDART00000145670
ENSDART00000146600 |
ctdsplb
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like b |
| chr5_-_38342992 | 0.62 |
ENSDART00000140337
|
mink1
|
misshapen-like kinase 1 |
| chr21_-_22693730 | 0.62 |
ENSDART00000158342
|
gig2d
|
grass carp reovirus (GCRV)-induced gene 2d |
| chr17_-_722218 | 0.62 |
ENSDART00000160385
|
SLC25A29
|
solute carrier family 25 member 29 |
| chr3_-_52644737 | 0.61 |
ENSDART00000126180
|
si:dkey-210j14.3
|
si:dkey-210j14.3 |
| chr15_+_17100412 | 0.61 |
ENSDART00000154418
|
relb
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr11_-_11458208 | 0.61 |
ENSDART00000005485
|
krt93
|
keratin 93 |
| chr23_-_437467 | 0.61 |
ENSDART00000192106
|
tspan2b
|
tetraspanin 2b |
| chr13_-_27916439 | 0.60 |
ENSDART00000139081
ENSDART00000087097 |
ogfrl1
|
opioid growth factor receptor-like 1 |
| chr24_-_11127493 | 0.60 |
ENSDART00000144066
|
myrip
|
myosin VIIA and Rab interacting protein |
| chr14_+_49381828 | 0.60 |
ENSDART00000169441
|
tnip1
|
TNFAIP3 interacting protein 1 |
| chr9_-_23894392 | 0.59 |
ENSDART00000133417
|
asb18
|
ankyrin repeat and SOCS box containing 18 |
| chr20_-_20533865 | 0.59 |
ENSDART00000125039
|
six6b
|
SIX homeobox 6b |
| chr8_-_20230559 | 0.59 |
ENSDART00000193677
|
mllt1a
|
MLLT1, super elongation complex subunit a |
| chr21_-_44009169 | 0.59 |
ENSDART00000028960
|
ndufa2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2 |
| chr16_+_26766423 | 0.58 |
ENSDART00000048036
|
gem
|
GTP binding protein overexpressed in skeletal muscle |
| chr2_-_53592532 | 0.58 |
ENSDART00000184066
|
ccl25a
|
chemokine (C-C motif) ligand 25a |
| chr21_-_3201027 | 0.58 |
ENSDART00000161256
|
zbtb7c
|
zinc finger and BTB domain containing 7C |
| chr23_+_3591690 | 0.58 |
ENSDART00000180822
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr8_+_25302172 | 0.58 |
ENSDART00000046182
ENSDART00000145316 |
gstm.3
|
glutathione S-transferase mu tandem duplicate 3 |
| chr7_+_39758714 | 0.57 |
ENSDART00000111278
|
sorcs2
|
sortilin-related VPS10 domain containing receptor 2 |
| chr1_+_41690402 | 0.57 |
ENSDART00000177298
|
fbxo41
|
F-box protein 41 |
| chr16_+_54691594 | 0.57 |
ENSDART00000168991
|
CABZ01080434.1
|
|
| chr16_-_22225295 | 0.57 |
ENSDART00000163519
|
LO017682.1
|
|
| chr5_+_9382301 | 0.57 |
ENSDART00000124017
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr24_+_29449690 | 0.56 |
ENSDART00000105743
ENSDART00000193556 ENSDART00000145816 |
ntng1a
|
netrin g1a |
| chr9_+_41156287 | 0.55 |
ENSDART00000189195
ENSDART00000186270 |
stat4
|
signal transducer and activator of transcription 4 |
| chr5_+_30608057 | 0.55 |
ENSDART00000133583
|
si:ch211-117m20.4
|
si:ch211-117m20.4 |
| chr9_-_32604414 | 0.55 |
ENSDART00000088876
ENSDART00000166502 |
satb2
|
SATB homeobox 2 |
| chr11_+_40544269 | 0.55 |
ENSDART00000175424
ENSDART00000184673 |
mmel1
|
membrane metallo-endopeptidase-like 1 |
| chr15_-_1962326 | 0.55 |
ENSDART00000179669
|
dock10
|
dedicator of cytokinesis 10 |
| chr20_+_37383034 | 0.55 |
ENSDART00000134997
ENSDART00000145927 |
adgrg6
|
adhesion G protein-coupled receptor G6 |
| chr14_-_1646360 | 0.54 |
ENSDART00000186528
|
LO018106.2
|
|
| chr3_+_55131289 | 0.53 |
ENSDART00000111585
|
AL935210.1
|
|
| chr18_-_6460102 | 0.53 |
ENSDART00000137037
|
iqsec3b
|
IQ motif and Sec7 domain 3b |
| chr6_-_442163 | 0.53 |
ENSDART00000163564
ENSDART00000189134 ENSDART00000169789 |
grap2b
|
GRB2-related adaptor protein 2b |
| chr22_-_31517300 | 0.52 |
ENSDART00000164799
|
slc6a6b
|
solute carrier family 6 (neurotransmitter transporter), member 6b |
| chr19_-_46088429 | 0.52 |
ENSDART00000161385
|
ptdss1b
|
phosphatidylserine synthase 1b |
| chr8_+_53452681 | 0.52 |
ENSDART00000166705
|
cacna1db
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, b |
| chr16_-_11798994 | 0.51 |
ENSDART00000135408
|
cnfn
|
cornifelin |
| chr3_+_13190012 | 0.51 |
ENSDART00000179747
ENSDART00000109876 ENSDART00000124824 ENSDART00000130261 |
sun1
|
Sad1 and UNC84 domain containing 1 |
| chr1_+_31113951 | 0.51 |
ENSDART00000129362
|
eef1a1b
|
eukaryotic translation elongation factor 1 alpha 1b |
| chr25_-_1333676 | 0.51 |
ENSDART00000183182
ENSDART00000188907 |
cln6b
|
CLN6, transmembrane ER protein b |
| chr16_+_23403602 | 0.50 |
ENSDART00000159848
|
s100w
|
S100 calcium binding protein W |
| chr12_+_22576404 | 0.50 |
ENSDART00000172053
|
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr18_+_2563756 | 0.50 |
ENSDART00000164147
|
p2ry2.1
|
purinergic receptor P2Y2, tandem duplicate 1 |
| chr20_-_45040916 | 0.49 |
ENSDART00000190001
|
klhl29
|
kelch-like family member 29 |
| chr11_-_396724 | 0.49 |
ENSDART00000184375
|
CR855375.3
|
|
| chr13_+_30692669 | 0.49 |
ENSDART00000187818
|
CR762483.1
|
|
| chr14_-_2267515 | 0.49 |
ENSDART00000180988
ENSDART00000130712 |
pcdh2aa15
|
protocadherin 2 alpha a 15 |
| chr7_-_15413019 | 0.48 |
ENSDART00000172147
|
si:ch211-106j24.1
|
si:ch211-106j24.1 |
| chr6_+_49771626 | 0.48 |
ENSDART00000134207
|
ctsz
|
cathepsin Z |
| chr12_+_41420355 | 0.48 |
ENSDART00000158811
|
epc1a
|
enhancer of polycomb homolog 1 (Drosophila) a |
| chr15_+_14109652 | 0.48 |
ENSDART00000156370
|
clca1
|
chloride channel accessory 1 |
| chr19_-_42249597 | 0.48 |
ENSDART00000184762
|
CR936363.1
|
|
| chr15_+_42235449 | 0.47 |
ENSDART00000114801
ENSDART00000182053 |
SGPP2
|
sphingosine-1-phosphate phosphatase 2 |
| chr12_+_5081759 | 0.47 |
ENSDART00000164178
|
prrt2
|
proline-rich transmembrane protein 2 |
| chr12_-_3903886 | 0.47 |
ENSDART00000184214
ENSDART00000041082 |
gdpd3b
|
glycerophosphodiester phosphodiesterase domain containing 3b |
| chr7_+_57677120 | 0.47 |
ENSDART00000110623
|
arsj
|
arylsulfatase family, member J |
| chr22_-_6801876 | 0.47 |
ENSDART00000135726
|
si:ch1073-188e1.1
|
si:ch1073-188e1.1 |
| chr22_-_34591295 | 0.47 |
ENSDART00000044429
|
rnf123
|
ring finger protein 123 |
| chr16_+_32995882 | 0.47 |
ENSDART00000170157
|
prss35
|
protease, serine, 35 |
| chr11_-_36765376 | 0.46 |
ENSDART00000163210
|
cacna2d3
|
calcium channel, voltage dependent, alpha2/delta subunit 3 |
| chr5_+_58996765 | 0.45 |
ENSDART00000062175
|
derl2
|
derlin 2 |
| chr25_+_8955530 | 0.44 |
ENSDART00000156444
|
si:ch211-256a21.4
|
si:ch211-256a21.4 |
| chr5_-_41841675 | 0.44 |
ENSDART00000141683
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr4_+_7888047 | 0.44 |
ENSDART00000104676
|
camk1da
|
calcium/calmodulin-dependent protein kinase 1Da |
| chr1_-_52431220 | 0.44 |
ENSDART00000111256
|
zgc:194101
|
zgc:194101 |
| chr23_-_43609595 | 0.44 |
ENSDART00000172222
|
CABZ01117603.1
|
|
| chr14_-_2209742 | 0.44 |
ENSDART00000054889
|
pcdh2ab5
|
protocadherin 2 alpha b 5 |
| chr18_+_27738349 | 0.43 |
ENSDART00000187816
|
tspan18b
|
tetraspanin 18b |
| chr4_-_33525219 | 0.43 |
ENSDART00000150491
|
CT027801.2
|
|
| chr12_-_25217217 | 0.43 |
ENSDART00000152931
|
kcng3
|
potassium voltage-gated channel, subfamily G, member 3 |
| chr25_-_13188214 | 0.42 |
ENSDART00000187298
|
si:ch211-147m6.1
|
si:ch211-147m6.1 |
| chr5_+_51909740 | 0.42 |
ENSDART00000162541
|
thbs4a
|
thrombospondin 4a |
| chr10_-_9115383 | 0.42 |
ENSDART00000139324
|
si:dkeyp-41f9.3
|
si:dkeyp-41f9.3 |
| chr3_-_40976288 | 0.42 |
ENSDART00000193553
|
cyp3c1
|
cytochrome P450, family 3, subfamily c, polypeptide 1 |
| chr19_-_307246 | 0.41 |
ENSDART00000145916
|
lingo4a
|
leucine rich repeat and Ig domain containing 4a |
| chr25_+_35706493 | 0.41 |
ENSDART00000176741
|
kif21a
|
kinesin family member 21A |
| chr6_+_49771372 | 0.41 |
ENSDART00000063251
|
ctsz
|
cathepsin Z |
| chr8_+_25629722 | 0.40 |
ENSDART00000026807
|
wasa
|
Wiskott-Aldrich syndrome (eczema-thrombocytopenia) a |
| chr17_-_7733037 | 0.40 |
ENSDART00000064657
|
stx11a
|
syntaxin 11a |
| chr8_-_5220125 | 0.40 |
ENSDART00000035676
|
bnip3la
|
BCL2 interacting protein 3 like a |
| chr22_-_16180467 | 0.40 |
ENSDART00000171331
ENSDART00000185607 |
vcam1b
|
vascular cell adhesion molecule 1b |
| chr10_-_29120515 | 0.40 |
ENSDART00000162016
ENSDART00000149140 |
zpld1a
|
zona pellucida-like domain containing 1a |
Network of associatons between targets according to the STRING database.
First level regulatory network of nfkb1_rela
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.5 | 1.4 | GO:0002468 | dendritic cell antigen processing and presentation(GO:0002468) |
| 0.4 | 1.3 | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 0.4 | 1.3 | GO:0035971 | peptidyl-histidine dephosphorylation(GO:0035971) |
| 0.4 | 1.3 | GO:0030857 | negative regulation of epithelial cell differentiation(GO:0030857) |
| 0.4 | 1.5 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.3 | 2.2 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.3 | 3.9 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.2 | 0.7 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.2 | 0.9 | GO:0015677 | copper ion import(GO:0015677) |
| 0.2 | 1.0 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.2 | 0.8 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.2 | 0.6 | GO:0072679 | thymocyte migration(GO:0072679) |
| 0.2 | 0.5 | GO:0060879 | peripheral nervous system myelin formation(GO:0032290) semicircular canal fusion(GO:0060879) |
| 0.2 | 0.9 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 2.5 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 1.2 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 0.4 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) |
| 0.1 | 0.5 | GO:0070255 | regulation of mucus secretion(GO:0070255) positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 0.6 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 1.6 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.1 | 0.5 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 | 0.3 | GO:0003097 | renal water homeostasis(GO:0003091) renal water transport(GO:0003097) |
| 0.1 | 0.7 | GO:0033292 | T-tubule organization(GO:0033292) |
| 0.1 | 0.8 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.1 | 1.5 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 2.0 | GO:0008345 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.1 | 1.2 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 0.2 | GO:0045191 | response to protozoan(GO:0001562) TRIF-dependent toll-like receptor signaling pathway(GO:0035666) defense response to protozoan(GO:0042832) regulation of isotype switching(GO:0045191) positive regulation of isotype switching(GO:0045830) |
| 0.1 | 1.3 | GO:0006584 | catecholamine metabolic process(GO:0006584) catechol-containing compound metabolic process(GO:0009712) |
| 0.1 | 0.9 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.1 | 0.2 | GO:0060945 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) Schwann cell migration(GO:0036135) cardiac neuron differentiation(GO:0060945) cardiac neuron development(GO:0060959) |
| 0.1 | 1.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.7 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.1 | 0.6 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.1 | 0.2 | GO:0060547 | negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.1 | 0.6 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.1 | 0.9 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.1 | 1.4 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 2.4 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 0.2 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.0 | 0.4 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.5 | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902108) |
| 0.0 | 1.8 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.2 | GO:0035777 | pronephric distal tubule development(GO:0035777) |
| 0.0 | 0.4 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.2 | GO:0090134 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.0 | 0.4 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.0 | 0.7 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.4 | GO:0019883 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.3 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.1 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.2 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 2.1 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.5 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.2 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.3 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.1 | GO:0061400 | positive regulation of transcription from RNA polymerase II promoter in response to calcium ion(GO:0061400) |
| 0.0 | 0.2 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 1.0 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.4 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.4 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.7 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.2 | GO:0016119 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.0 | 1.1 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.0 | 0.2 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.4 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.0 | 0.9 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.9 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.5 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.1 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.0 | 0.5 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.1 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.4 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 0.1 | GO:0045649 | regulation of macrophage differentiation(GO:0045649) |
| 0.0 | 0.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.6 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.9 | GO:0097696 | JAK-STAT cascade(GO:0007259) STAT cascade(GO:0097696) |
| 0.0 | 0.3 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.6 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 0.5 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.0 | GO:0048016 | inositol phosphate-mediated signaling(GO:0048016) |
| 0.0 | 0.5 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0008207 | C21-steroid hormone metabolic process(GO:0008207) |
| 0.0 | 1.2 | GO:0048741 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.0 | 0.5 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 0.0 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.5 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.3 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.7 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.2 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.0 | 0.6 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.3 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.2 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.5 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 2.5 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.2 | 1.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 0.5 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.9 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.1 | 1.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 1.6 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 1.5 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.5 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.1 | 2.1 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.1 | 0.4 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 3.0 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 1.4 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 2.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 1.2 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.8 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0070743 | interleukin-12 complex(GO:0043514) interleukin-23 complex(GO:0070743) |
| 0.0 | 0.4 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 2.2 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.5 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 2.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.9 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.8 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 1.0 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.7 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.6 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.2 | GO:0032589 | neuron projection membrane(GO:0032589) dendrite membrane(GO:0032590) |
| 0.0 | 0.7 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.1 | GO:0034703 | cation channel complex(GO:0034703) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.3 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.5 | 1.6 | GO:0004031 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.3 | 1.3 | GO:0101006 | protein histidine phosphatase activity(GO:0101006) |
| 0.3 | 1.0 | GO:0031530 | gonadotropin hormone-releasing hormone activity(GO:0005183) gonadotropin-releasing hormone receptor binding(GO:0031530) |
| 0.3 | 1.2 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.3 | 1.5 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.2 | 0.9 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.2 | 1.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.2 | 1.6 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.2 | 0.5 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 1.3 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 1.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 1.5 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 2.1 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 2.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 2.1 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 0.3 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.1 | 1.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.7 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.1 | 0.2 | GO:0031704 | apelin receptor binding(GO:0031704) |
| 0.1 | 1.3 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 0.6 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.6 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 0.4 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 0.8 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 1.6 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.7 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.1 | 0.2 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.1 | 2.4 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.2 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 2.1 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.5 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.4 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.3 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.0 | 2.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.3 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.1 | GO:0019972 | interleukin-12 binding(GO:0019972) interleukin-12 alpha subunit binding(GO:0042164) |
| 0.0 | 0.5 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.2 | GO:0005549 | olfactory receptor activity(GO:0004984) odorant binding(GO:0005549) |
| 0.0 | 0.4 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.2 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.0 | 0.5 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 1.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.3 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.7 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.8 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0017020 | myosin phosphatase regulator activity(GO:0017020) |
| 0.0 | 3.0 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.5 | GO:1901682 | sulfur compound transmembrane transporter activity(GO:1901682) |
| 0.0 | 2.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 0.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 1.8 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.4 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.2 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.3 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 0.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 1.3 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.6 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.2 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.5 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.9 | GO:0019209 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.0 | 1.4 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.7 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 2.3 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.2 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.6 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.3 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.1 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.4 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.2 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.2 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.2 | 0.2 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 2.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 1.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.7 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.1 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 1.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.4 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.2 | 3.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 0.8 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 3.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 0.9 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 1.2 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.5 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 1.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.9 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |